IdentifyRegulator
IdentifyRegulator.RmdIntroduction
Since there are many publicly available ChIP-seq datasets, many tools have been developed to identify TFs from these data. Here, DEbPeak provide three methods to identify TFs from gene expression data:
- BART: only supports human and mouse gene symbols.
- ChEA3: only supports human and mouse gene symbols.
- TFEA.ChIP: only supports mouse.
While ARACNe-AP (another widely used tool) reguires hundreds of samples, it is not included in DEbPeak.
Example data
The data used here are from In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer:
- RNA-seq data: the sgCtrl vs sgFli1 RNA sequencing at D8 Cl13 p.i., the raw data are stored in GSE149838
There are also ATAC-seq datasets available, users can compare the results obatined from these methods with ATAC-seq data.
Load packages
# library
suppressWarnings(suppressMessages(library(tidyverse)))
suppressWarnings(suppressMessages(library(DEbPeak)))Differential expression analysis
DEbPeak has prepared the differential expression analysis results:
rna.diff.file <- system.file("extdata", "RA_RNA_diff.txt", package = "DEbPeak")
rna.diff <- read.table(file = rna.diff.file, header = TRUE, sep = "\t")Prepare gene sets
BART and ChEA3 require differentially expressed genes as input, TFEA.ChIP requires differentially expressed genes and non-differentially expressed genes (as background genes) as inputs.
# up-regulated genes
rna.up.sig.df = rna.diff %>%
dplyr::filter(padj < 0.05 & log2FoldChange > 0)
# non-differentially expressed genes
rna.contorl.df = rna.diff %>%
dplyr::filter(padj > 0.5 & abs(log2FoldChange) <0.25)While TFEA.ChIP only support human genes, we need to convert the mouse genes to their human orthologs.
# convert mouse gene symbol to human entrez id
Mouse2HumanEntrez <- function(x){
require("biomaRt")
human = biomaRt::useMart("ensembl", dataset = "hsapiens_gene_ensembl", host = "https://dec2021.archive.ensembl.org/")
mouse = biomaRt::useMart("ensembl", dataset = "mmusculus_gene_ensembl", host = "https://dec2021.archive.ensembl.org/")
m2h = biomaRt::getLDS(attributes = c("mgi_symbol"), filters = "mgi_symbol", values = x ,
mart = mouse, attributesL = c("entrezgene_id"), martL = human, uniqueRows=T)
m2h.ids <- unique(m2h[, 2])
return(m2h.ids)
}
# convert mouse gene symbol to human entrez id
rna.upsig.ids = Mouse2HumanEntrez(x = rownames(rna.up.sig.df))
write.table(x = rna.upsig.ids, file = "/home/songyabing/R/learn/tmp/DEbPeak/RNAinfer/ATACDE_padj0.05_lfc0.txt",
quote = F, row.names = F, col.names = F)
rna.contorl.ids = Mouse2HumanEntrez(x = rownames(rna.contorl.df))
write.table(x = rna.contorl.ids, file = "/home/songyabing/R/learn/tmp/DEbPeak/RNAinfer/ATACDE_padj0.5_lfc0.25_contorl.txt",
quote = F, row.names = F, col.names = F)The above codes are slow, we will load pre-run results:
rna.upsig.ids.df = read.table("/home/songyabing/R/learn/tmp/DEbPeak/RNAinfer/ATACDE_padj0.05_lfc0.txt")
rna.upsig.ids = rna.upsig.ids.df$V1
rna.contorl.ids.df = read.table("/home/songyabing/R/learn/tmp/DEbPeak/RNAinfer/ATACDE_padj0.5_lfc0.25_contorl.txt")
rna.contorl.ids = rna.contorl.ids.df$V1Identify TFs with ChEA3
chea3.res = InferRegulator(genes = rownames(rna.up.sig.df), method = "ChEA3", species = "Mouse")##
## 载入程辑包:'BiocGenerics'## The following objects are masked from 'package:lubridate':
##
## intersect, setdiff, union## The following objects are masked from 'package:dplyr':
##
## combine, intersect, setdiff, union## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs## The following objects are masked from 'package:base':
##
## anyDuplicated, append, as.data.frame, basename, cbind, colnames,
## dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
## grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
## order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
## rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
## union, unique, unsplit, which.max, which.min## Welcome to Bioconductor
##
## Vignettes contain introductory material; view with
## 'browseVignettes()'. To cite Bioconductor, see
## 'citation("Biobase")', and for packages 'citation("pkgname")'.##
## 载入程辑包:'S4Vectors'## The following objects are masked from 'package:lubridate':
##
## second, second<-## The following objects are masked from 'package:dplyr':
##
## first, rename## The following object is masked from 'package:tidyr':
##
## expand## The following object is masked from 'package:base':
##
## expand.grid##
## 载入程辑包:'IRanges'## The following object is masked from 'package:lubridate':
##
## %within%## The following objects are masked from 'package:dplyr':
##
## collapse, desc, slice## The following object is masked from 'package:purrr':
##
## reduce##
## 载入程辑包:'AnnotationDbi'## The following object is masked from 'package:dplyr':
##
## select##
head(chea3.res)## Query Name Rank TF Score
## 1 DEbPeak 1 ZNF367 5.0
## 2 DEbPeak 2 CENPA 5.0
## 3 DEbPeak 3 FOXM1 11.67
## 4 DEbPeak 4 E2F7 21.8
## 5 DEbPeak 5 MYBL2 34.0
## 6 DEbPeak 6 ZNF695 37.67
## Library
## 1 ARCHS4 Coexpression,11;Enrichr Queries,1;GTEx Coexpression,3
## 2 ARCHS4 Coexpression,2;GTEx Coexpression,8
## 3 Literature ChIP-seq,3;ARCHS4 Coexpression,58;ENCODE ChIP-seq,2;Enrichr Queries,4;ReMap ChIP-seq,1;GTEx Coexpression,2
## 4 Literature ChIP-seq,18;ARCHS4 Coexpression,3;Enrichr Queries,6;ReMap ChIP-seq,75;GTEx Coexpression,7
## 5 Literature ChIP-seq,23;ARCHS4 Coexpression,40;ENCODE ChIP-seq,68;Enrichr Queries,2;ReMap ChIP-seq,62;GTEx Coexpression,9
## 6 ARCHS4 Coexpression,92;Enrichr Queries,10;GTEx Coexpression,11
## Overlapping_Genes
## 1 PI4K2B,ERCC6L,ZWILCH,HJURP,SMC4,SMC2,EME1,STMN1,PCLAF,NUSAP1,KNTC1,FBXO5,TMPO,WDHD1,ESCO2,CDC25C,SGO1,CCNE2,CCNE1,TIMELESS,CD226,TLN1,PRR11,ANAPC1,ASF1B,FKBP5,DNMT1,CDCA2,SLC35D3,NCAPG,RAD51AP1,CCNB2,DSN1,ORC6,RACGAP1,FIGNL1,PMAIP1,ECT2,PLK4,NUP155,DDIAS,CDC6,NDC80,HNRNPL,POLA1,CDK6,NASP,CDK2,ANP32E,EZH2,ARHGAP11A,TOP2A,GPSM2,NCAPG2,HMGB2,BRCA1,KIF11,FOXM1,KIF15,XPO1,NUF2,PBK,TOPBP1,LBR,POLE,DLGAP5,CEP55,HELLS,CKAP2L,MMD,CKAP2,HAUS6,CIP2A,FOS,CCNA2,ASPM,RBL1,DBF4,ESPL1,MCM4,MCM6,DTL,MTPN,PCNA,NUFIP2,UHRF1,SRSF1,TTK,TYMS,KCTD20,USP1,BUB1,E2F7,E2F8,RRM1,RRM2,CDKN2C,ATAD5,ATAD2,DONSON,DEK,CENPF,WEE1,CENPH,PRC1,RFWD3,RPA3,POLE2,CENPK,CENPN,NCAPD2,NCAPD3,MAD2L1,SPC25,CDKN3
## 2 ARHGAP11A,TOP2A,PI4K2B,ERCC6L,ARL6IP1,NCAPG2,HJURP,HMGB2,BRCA1,KIF11,FOXM1,SMC4,SMC2,KIF15,XPO1,EME1,NUF2,PCLAF,NUSAP1,PBK,KNTC1,FBXO5,TOPBP1,HMGN2,DLGAP5,CEP55,TMPO,WDHD1,HELLS,CKAP2L,CAD,CKAP2,NIPA2,ESCO2,CIP2A,CDC25C,CKAP5,SGO1,CCNA2,ASPM,RBL1,DBF4,ESPL1,CCNE1,TIMELESS,G2E3,MCM4,MCM6,PRR11,DTL,ANAPC1,ASF1B,PCNA,CDCA2,UHRF1,SRSF1,NCAPG,TTK,TYMS,RAD51AP1,CCNB2,ORC6,RACGAP1,FIGNL1,USP1,PMAIP1,ECT2,BUB1,E2F7,E2F8,PLK4,RRM1,RRM2,CDKN2C,ATAD5,DDIAS,ATAD2,CDC6,DEK,NDC80,HNRNPL,CENPF,CENPH,PRC1,RPA3,POLE2,CENPK,HNRNPD,NCAPD2,CENPN,NCAPD3,EZH2,MAD2L1,SPC25,CDKN3
## 3 EIF4A1,ERCC6L,ARL6IP1,ZWILCH,HJURP,SMC6,SMC4,SMC2,MT-ND1,LGALS3,EME1,STMN1,PCLAF,NUSAP1,PIM1,KNTC1,FBXO5,CCNL1,TMPO,EPHB3,WDHD1,TPM4,LIG1,CMC2,DYRK1A,P3H4,ESCO2,CDC25C,NEAT1,GEM,SGO1,KCTD9,CCNE2,CCNE1,DNAJB11,TIMELESS,FAR1,FAM76B,MATR3,NUP98,SNRNP200,SRSF6,PRR11,ASF1B,SLBP,DNMT1,DDX5,CDCA2,NOTCH1,TSHZ1,NCAPG,STK4,NUP160,HSP90B1,RAD51AP1,CCNB2,VCPIP1,DSN1,ORC6,RACGAP1,FIGNL1,ERI2,CEP72,PMAIP1,TRDC,ECT2,SLC38A2,S100A10,SLAMF1,PLK4,OSBPL9,CDK19,H3F3B,DDIAS,RDX,CDC6,NDC80,HNRNPL,EHD1,MOB1A,POLA1,NASP,CNOT1,CDK2,ANP32E,HNRNPD,MDM4,EZH2,ATR,ARHGAP11A,TOP2A,CEP57,GPSM2,PHTF2,NCAPG2,CLTC,HP1BP3,HMGB2,BRCA1,KIF11,LAMC1,FOXM1,KIF15,SEPT7,XPO1,SFR1,NUF2,PBK,TOPBP1,HIF1AN,CFAP43,LBR,POLE,DLGAP5,CEP55,POLG,HELLS,PLEKHG2,PCYT1A,ANXA2,CKAP2L,CAD,CKAP2,EMP1,HAUS6,CIP2A,FOS,CKAP5,CCNA2,ASPM,PTP4A2,DBF4,RBL1,ESPL1,ZADH2,DMC1,G2E3,MCM4,MCM6,DTL,CD44,PPP1R15B,PCNA,UHRF1,ATL2,SRSF1,MIS12,NEDD9,TTK,LIN9,TYMS,CLCN3,MALT1,U2AF2,PPP1R7,USP1,FLNA,BRD8,BUB1,SRSF10,E2F7,E2F8,RBM39,RRM1,RRM2,CDKN2C,ATAD5,CSNK1A1,NFYB,SEMA4B,ATAD2,RSRC2,PTPN13,PNRC2,ASXL1,PDP1,CENPF,KLF6,WEE1,CENPH,PRC1,RFWD3,POLE2,CENPK,NCAPD2,CENPN,NCAPD3,CDKN3,MAD2L1,SPC25,TJP2
## 4 PI4K2B,EIF4A1,ERCC6L,UBE3C,ZWILCH,HJURP,SMC4,SMC2,EME1,STMN1,PCLAF,NUSAP1,PIM1,KNTC1,FBXO5,TMPO,WDHD1,TPM4,LIG1,DYRK1A,P3H4,ESCO2,CDC25C,NEAT1,SGO1,CCNE2,CCNE1,TIMELESS,CD226,PRR11,ANAPC1,ASF1B,SLBP,DNMT1,DDX5,CDCA2,NOTCH1,NCAPG,PDS5A,RAD51AP1,CCNB2,DSN1,ORC6,RACGAP1,FIGNL1,ERI2,PMAIP1,ECT2,SLC38A2,SLAMF1,PLK4,H3F3B,NUP155,DDIAS,BICRAL,PTCH1,CDC6,NDC80,EHD1,HNRNPL,POLA1,CDK6,CDK2,HNRNPD,MDM4,NFE2L1,EZH2,ARHGAP11A,TOP2A,GPSM2,CLIC4,NCAPG2,CLTC,HMGB2,BRCA1,KIF11,CTDSPL2,FOXM1,KIF15,SYNCRIP,TUBA1A,XPO1,GLIPR2,NUF2,PBK,TOPBP1,POLE,DLGAP5,CEP55,HELLS,PLEKHG2,ANXA2,RFC1,CKAP2L,CAD,CKAP2,EMP1,HAUS6,CIP2A,FOS,CKAP5,CCNA2,ASPM,PTP4A2,DBF4,RBL1,ESPL1,MCM4,MYH9,MCM6,ANGPTL4,GAS6,DTL,PCNA,UHRF1,SRSF1,TTK,TYMS,U2AF2,MXI1,USP1,FLNA,BUB1,SRSF10,E2F8,P2RY10,RRM1,RRM2,CDKN2C,ATAD5,USP9X,ATAD2,RSRC2,DEK,CENPF,KLF6,WEE1,CENPH,PRC1,RPA3,POLE2,CENPK,NCAPD2,CENPN,NCAPD3,MAD2L1,SPC25,CDKN3
## 5 USP6NL,EIF4A1,ARL6IP1,SMC5,SMC6,SMC4,SMC2,MT-ND1,GPR171,STMN1,PCLAF,FBXO5,CCNL1,XYLB,TMPO,WDHD1,AQR,WSB1,NUP210,ESCO2,MED7,WDR82,SUB1,TMEM126A,PRR11,CGGBP1,ASF1B,CDCA2,CSTF3,NCAPG,ANAPC10,UBE2J1,DSN1,RACGAP1,ERI1,FIGNL1,ERI2,SLAMF1,PLK4,CDK19,H3F3B,DDIAS,CDC6,EHD1,HNRNPL,POLA1,ID2,ANP32E,HNRNPD,EZH2,DCP1B,TOP2A,CEP57,TRAM1,NCAPG2,CLTC,AP4E1,BRCA1,KIF11,KIF15,XPO1,TUBA1A,RARS,NUF2,PBK,DHX15,ARIH1,JAK2,DLGAP5,CEP55,HELLS,PCYT1A,APOBEC2,ANXA2,GSTO1,MMD,IFNGR1,FOS,FAM126A,HNRNPH1,FRG1,G2E3,CD48,ANGPTL4,PCNA,ATL2,NEDD9,ABRACL,CAPG,TYMS,TMEM168,EPCAM,PPP1R7,USP1,BRD8,SRSF10,P2RY10,NQO2,CDKN2C,ATAD5,YTHDF3,C1GALT1,ATAD2,RPA2,RSRC2,PLXDC1,VMP1,CENPF,CENPH,RFWD3,RPA3,CAPZA1,CENPK,CAPZA2,RLIM,NCAPD2,CENPN,NCAPD3,MAD2L1,SPC25,CDKN3,ERCC6L,ZWILCH,HJURP,EME1,NUSAP1,PIM1,KNTC1,PDIA3,CAST,LIG1,TPM4,PRPF4B,ATP11B,CDC25C,NEAT1,SGO1,KCTD9,CCNE2,CCNE1,TIMELESS,MATR3,SNRNP200,ANAPC1,DNMT1,ABHD2,COPB1,PSEN1,LIN7C,KRCC1,NUP160,HSP90B1,RAD51AP1,CCNB2,ORC6,ORC3,ECT2,SLC38A2,NUP155,PTCH1,CWC15,NDC80,CLK1,H1F0,CDK6,NASP,UBLCP1,CDK2,DNAJC10,TUBGCP6,MDM4,ARHGAP11A,GPSM2,RBM25,NGLY1,HP1BP3,CHD7,HMGB2,FOXM1,CASP3,PDE4A,TOPBP1,HIF1AN,CFAP43,LBR,POLE,PLEKHG2,CKAP2L,CAD,CKAP2,CIP2A,GPCPD1,YWHAZ,CKAP5,PTBP3,CCNA2,ASPM,DBF4,RBL1,ESPL1,MCM4,MCM6,DOCK2,DTL,CHMP5,PPP1R15B,MTPN,HLF,UHRF2,UHRF1,SRSF1,TTK,ATP1A1,CLCN3,RNFT1,CDC42,FAM168B,TNKS2,U2AF2,VPS54,MAPK6,BUB1,E2F7,E2F8,RBM39,RRM1,RRM2,TMEM30A,SEMA4B,KLF3,PNRC2,ASXL1,KLF6,WEE1,MYO1C,PRC1,POLE2,OCIAD1,SUGT1,RBMX,SSBP3
## 6 PI4K2B,ERCC6L,HJURP,SMC4,SMC2,EME1,STMN1,PCLAF,NUSAP1,KNTC1,FBXO5,TMPO,WDHD1,NUP210,KHNYN,ESCO2,CDC25C,SGO1,CCNE2,CCNE1,TIMELESS,SRSF6,TLN1,PRR11,ANAPC1,ASF1B,CDCA2,NCAPG,RAD51AP1,CCNB2,ABR,DSN1,ORC6,RACGAP1,FIGNL1,PMAIP1,ECT2,SLAMF1,PLK4,DDIAS,CDC6,NDC80,NASP,NFE2L1,EZH2,ARHGAP11A,TOP2A,GPSM2,NCAPG2,HMGB2,BRCA1,KIF11,FOXM1,KIF15,NUF2,PBK,TOPBP1,DLGAP5,CEP55,HELLS,CKAP2L,CKAP2,CIP2A,FOS,CCNA2,ASPM,DBF4,RBL1,ESPL1,G2E3,MCM4,MYH9,CD48,MCM6,DTL,PCNA,UHRF1,TTK,LIN9,TYMS,ADD1,EPCAM,U2AF2,BUB1,SRSF10,E2F7,E2F8,P2RY10,RRM1,RRM2,SSB,ATAD5,ATAD2,DONSON,MYO9B,CENPF,CENPH,PRC1,RPA3,POLE2,CENPK,NCAPD2,CENPN,SUGT1,MAD2L1,SPC25,CDKN3Visualize the results:
VizRegulator(infer.res = chea3.res, method = "ChEA3", label.gene = c("RUNX1", "RUNX2", "RUNX3"))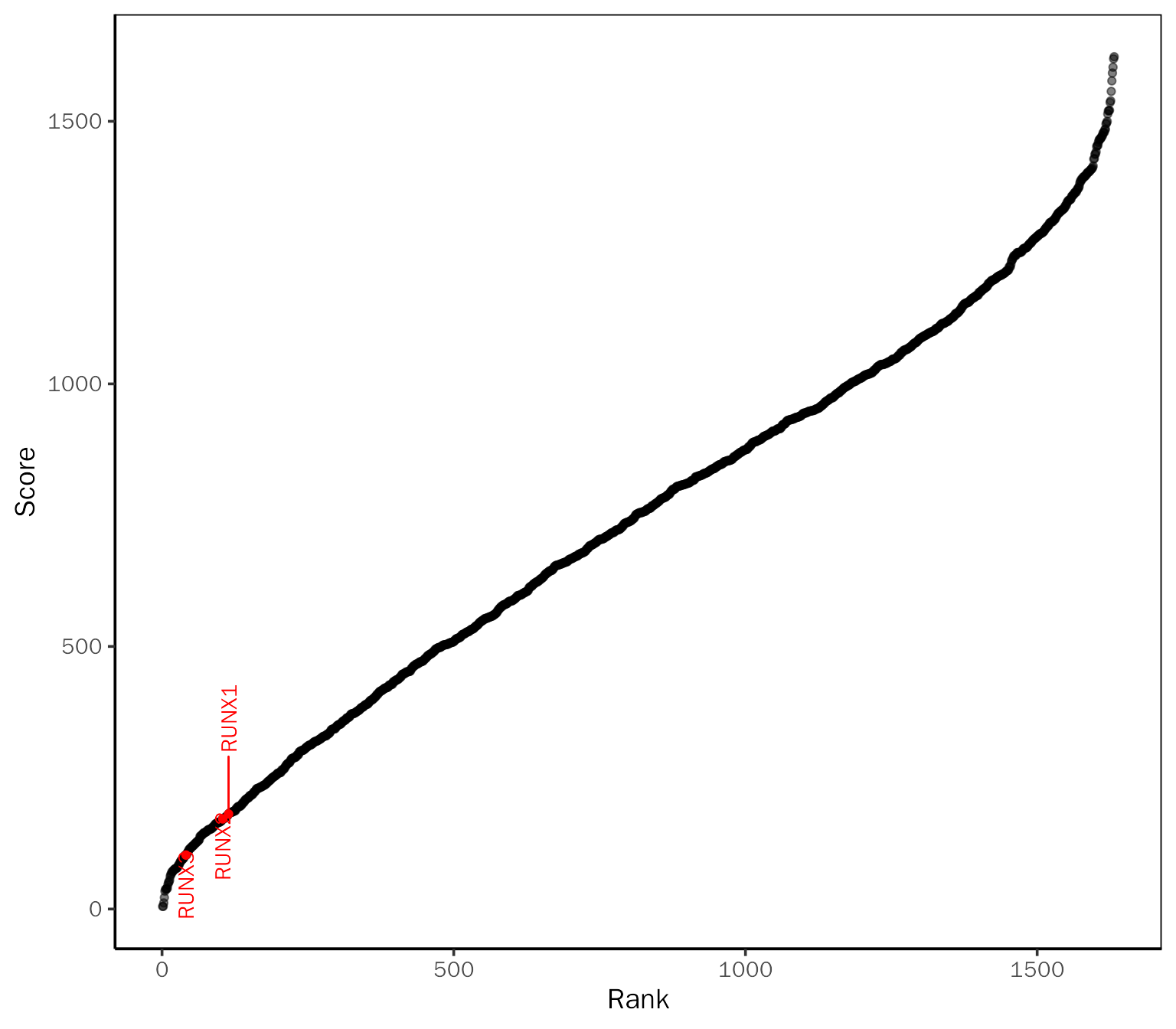
Identify TFs with BART2
bart2.res = InferRegulator(genes = rownames(rna.up.sig.df), method = "BART2",
species = "Mouse", bart2.path = "~/anaconda3/envs/bart2/bin/bart2")## Running BART2: ~/anaconda3/envs/bart2/bin/bart2 geneset -i /tmp/RtmpUzymaP/bart2_genes_328a3251fc286.txt -s mm10 --outdir /tmp/RtmpUzymaP/bart2 -o DEbPeak
head(bart2.res)## TF statistic pvalue zscore max_auc re_rank irwin_hall_pvalue
## 1 STAT2 4.176 1.484e-05 1.526 0.944 0.027 8.727e-05
## 2 FOXP3 5.055 2.147e-07 2.254 0.905 0.040 2.945e-04
## 3 STAT4 2.848 2.200e-03 2.225 0.918 0.046 4.421e-04
## 4 IRF4 7.796 3.183e-15 2.533 0.888 0.051 5.904e-04
## 5 STAT1 6.076 6.157e-10 0.943 0.933 0.053 6.536e-04
## 6 ETS1 4.170 1.520e-05 1.086 0.923 0.054 6.982e-04Visualize the results:
VizRegulator(infer.res = bart2.res, method = "BART2", label.gene = c("RUNX1", "RUNX2", "RUNX3"))
Identify TFs with TFEA.ChIP
tfea.res = InferRegulator(genes = rna.upsig.ids, control.genes = rna.contorl.ids, method = "TFEA.ChIP")
head(tfea.res)## TF ES arg.ES pVal numberOfChIPs
## MEF2C MEF2C 0.98584 16 0.02 1
## ATF2 ATF2 0.98339 6 0.00 2
## NFATC1 NFATC1 0.98206 20 0.05 1
## TBX21 TBX21 0.97545 27 0.06 1
## PRDM1 PRDM1 0.96317 40 0.06 1
## EBF1 EBF1 0.95736 44 0.03 2Visualize the results:
VizRegulator(infer.res = tfea.res, method = "TFEA.ChIP", label.gene = c("RUNX1", "RUNX2", "RUNX3"))
Session info
## R version 4.0.3 (2020-10-10)
## Platform: x86_64-conda-linux-gnu (64-bit)
## Running under: CentOS Linux 7 (Core)
##
## Matrix products: default
## BLAS/LAPACK: /home/softwares/anaconda3/envs/r4.0/lib/libopenblasp-r0.3.12.so
##
## locale:
## [1] LC_CTYPE=zh_CN.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=zh_CN.UTF-8 LC_COLLATE=zh_CN.UTF-8
## [5] LC_MONETARY=zh_CN.UTF-8 LC_MESSAGES=zh_CN.UTF-8
## [7] LC_PAPER=zh_CN.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=zh_CN.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] org.Hs.eg.db_3.12.0 org.Mm.eg.db_3.12.0 AnnotationDbi_1.52.0
## [4] IRanges_2.24.1 S4Vectors_0.28.1 Biobase_2.50.0
## [7] BiocGenerics_0.42.0 DEbPeak_1.4.0 lubridate_1.9.2
## [10] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.2
## [13] purrr_1.0.1 readr_2.1.4 tidyr_1.3.0
## [16] tibble_3.2.1 ggplot2_3.4.2 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] rsvd_1.0.3
## [2] ggvenn_0.1.9
## [3] apeglm_1.12.0
## [4] Rsamtools_2.6.0
## [5] rsvg_2.1
## [6] foreach_1.5.1
## [7] rprojroot_2.0.2
## [8] crayon_1.4.1
## [9] V8_3.4.2
## [10] MASS_7.3-58
## [11] nlme_3.1-152
## [12] backports_1.2.1
## [13] sva_3.38.0
## [14] GOSemSim_2.25.0
## [15] rlang_1.1.0
## [16] XVector_0.30.0
## [17] readxl_1.4.2
## [18] irlba_2.3.5
## [19] limma_3.46.0
## [20] GOstats_2.56.0
## [21] BiocParallel_1.24.1
## [22] rjson_0.2.20
## [23] bit64_4.0.5
## [24] glue_1.6.2
## [25] DiffBind_3.0.15
## [26] mixsqp_0.3-43
## [27] pheatmap_1.0.12
## [28] parallel_4.0.3
## [29] DEFormats_1.18.0
## [30] base64url_1.4
## [31] tcltk_4.0.3
## [32] DOSE_3.23.2
## [33] haven_2.5.2
## [34] tidyselect_1.2.0
## [35] SummarizedExperiment_1.20.0
## [36] rio_0.5.27
## [37] XML_3.99-0.6
## [38] ggpubr_0.4.0
## [39] GenomicAlignments_1.26.0
## [40] xtable_1.8-4
## [41] ggnetwork_0.5.12
## [42] magrittr_2.0.3
## [43] evaluate_0.14
## [44] cli_3.6.1
## [45] zlibbioc_1.36.0
## [46] hwriter_1.3.2
## [47] rstudioapi_0.14
## [48] bslib_0.3.1
## [49] GreyListChIP_1.22.0
## [50] fastmatch_1.1-3
## [51] BiocSingular_1.6.0
## [52] xfun_0.30
## [53] askpass_1.1
## [54] clue_0.3-59
## [55] gson_0.0.9
## [56] cluster_2.1.1
## [57] caTools_1.18.2
## [58] tidygraph_1.2.0
## [59] ggrepel_0.9.1
## [60] Biostrings_2.58.0
## [61] png_0.1-7
## [62] withr_2.5.0
## [63] bitops_1.0-6
## [64] ggforce_0.3.3
## [65] cellranger_1.1.0
## [66] RBGL_1.66.0
## [67] plyr_1.8.6
## [68] GSEABase_1.52.1
## [69] pcaPP_2.0-1
## [70] dqrng_0.2.1
## [71] coda_0.19-4
## [72] pillar_1.9.0
## [73] gplots_3.1.1
## [74] GlobalOptions_0.1.2
## [75] cachem_1.0.4
## [76] GenomicFeatures_1.42.2
## [77] fs_1.5.0
## [78] GetoptLong_1.0.5
## [79] clusterProfiler_4.7.1
## [80] DelayedMatrixStats_1.12.3
## [81] vctrs_0.6.2
## [82] generics_0.1.0
## [83] plot3D_1.4
## [84] tools_4.0.3
## [85] foreign_0.8-81
## [86] NOISeq_2.34.0
## [87] munsell_0.5.0
## [88] tweenr_1.0.2
## [89] fgsea_1.16.0
## [90] DelayedArray_0.16.3
## [91] abind_1.4-5
## [92] fastmap_1.1.0
## [93] compiler_4.0.3
## [94] rtracklayer_1.50.0
## [95] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
## [96] GenomeInfoDbData_1.2.4
## [97] gridExtra_2.3
## [98] edgeR_3.32.1
## [99] lattice_0.20-45
## [100] ggnewscale_0.4.7
## [101] AnnotationForge_1.32.0
## [102] utf8_1.2.1
## [103] BiocFileCache_1.14.0
## [104] jsonlite_1.8.4
## [105] scales_1.2.1
## [106] graph_1.68.0
## [107] carData_3.0-4
## [108] sparseMatrixStats_1.2.1
## [109] TFEA.ChIP_1.10.0
## [110] genefilter_1.72.1
## [111] car_3.0-11
## [112] doParallel_1.0.16
## [113] latticeExtra_0.6-29
## [114] R.utils_2.12.0
## [115] brew_1.0-6
## [116] checkmate_2.0.0
## [117] rmarkdown_2.14
## [118] openxlsx_4.2.3
## [119] pkgdown_1.6.1
## [120] cowplot_1.1.1
## [121] textshaping_0.3.6
## [122] downloader_0.4
## [123] BSgenome_1.58.0
## [124] igraph_1.4.99.9024
## [125] survival_3.2-10
## [126] numDeriv_2016.8-1.1
## [127] yaml_2.2.1
## [128] plotrix_3.8-2
## [129] systemfonts_1.0.4
## [130] ashr_2.2-47
## [131] SQUAREM_2021.1
## [132] htmltools_0.5.2
## [133] memoise_2.0.0
## [134] VariantAnnotation_1.36.0
## [135] locfit_1.5-9.4
## [136] graphlayouts_0.7.1
## [137] batchtools_0.9.15
## [138] PCAtools_2.2.0
## [139] viridisLite_0.4.0
## [140] rrcov_1.7-0
## [141] digest_0.6.27
## [142] assertthat_0.2.1
## [143] rappdirs_0.3.3
## [144] emdbook_1.3.12
## [145] RSQLite_2.2.5
## [146] amap_0.8-18
## [147] yulab.utils_0.0.4
## [148] debugme_1.1.0
## [149] misc3d_0.9-1
## [150] data.table_1.14.2
## [151] blob_1.2.1
## [152] R.oo_1.24.0
## [153] ragg_0.4.0
## [154] labeling_0.4.2
## [155] splines_4.0.3
## [156] ggupset_0.3.0
## [157] RCurl_1.98-1.3
## [158] broom_1.0.4
## [159] hms_1.1.3
## [160] colorspace_2.0-0
## [161] BiocManager_1.30.16
## [162] GenomicRanges_1.42.0
## [163] shape_1.4.6
## [164] sass_0.4.1
## [165] GEOquery_2.58.0
## [166] Rcpp_1.0.9
## [167] mvtnorm_1.1-2
## [168] circlize_0.4.15
## [169] enrichplot_1.10.2
## [170] fansi_0.4.2
## [171] tzdb_0.3.0
## [172] truncnorm_1.0-8
## [173] ChIPseeker_1.33.0.900
## [174] R6_2.5.0
## [175] grid_4.0.3
## [176] lifecycle_1.0.3
## [177] ShortRead_1.48.0
## [178] zip_2.1.1
## [179] curl_4.3
## [180] ggsignif_0.6.3
## [181] jquerylib_0.1.3
## [182] robustbase_0.95-0
## [183] DO.db_2.9
## [184] Matrix_1.5-4
## [185] qvalue_2.22.0
## [186] desc_1.3.0
## [187] RColorBrewer_1.1-2
## [188] iterators_1.0.13
## [189] DOT_0.1
## [190] ggpie_0.2.5
## [191] beachmat_2.6.4
## [192] polyclip_1.10-0
## [193] biomaRt_2.46.3
## [194] shadowtext_0.0.9
## [195] timechange_0.2.0
## [196] gridGraphics_0.5-1
## [197] mgcv_1.8-34
## [198] ComplexHeatmap_2.13.1
## [199] openssl_1.4.3
## [200] patchwork_1.0.0
## [201] bdsmatrix_1.3-4
## [202] codetools_0.2-18
## [203] matrixStats_0.58.0
## [204] invgamma_1.1
## [205] GO.db_3.12.1
## [206] gtools_3.8.2
## [207] prettyunits_1.1.1
## [208] dbplyr_2.3.2
## [209] R.methodsS3_1.8.1
## [210] GenomeInfoDb_1.26.7
## [211] gtable_0.3.0
## [212] DBI_1.1.1
## [213] highr_0.8
## [214] ggfun_0.0.6
## [215] httr_1.4.5
## [216] KernSmooth_2.23-18
## [217] stringi_1.5.3
## [218] progress_1.2.2
## [219] reshape2_1.4.4
## [220] farver_2.1.0
## [221] annotate_1.68.0
## [222] viridis_0.6.1
## [223] Rgraphviz_2.34.0
## [224] xml2_1.3.4
## [225] bbmle_1.0.24
## [226] systemPipeR_1.24.3
## [227] boot_1.3-28
## [228] geneplotter_1.68.0
## [229] ggplotify_0.1.0
## [230] Category_2.56.0
## [231] DEoptimR_1.0-11
## [232] DESeq2_1.30.1
## [233] bit_4.0.4
## [234] scatterpie_0.1.7
## [235] jpeg_0.1-8.1
## [236] MatrixGenerics_1.2.1
## [237] ggraph_2.0.5
## [238] pkgconfig_2.0.3
## [239] rstatix_0.7.0
## [240] knitr_1.37