DifferentialExpressionAnalysis
DifferentialExpressionAnalysis.RmdIntroduction
Here, we will perform differential expression analysis based on DESeq2 and edgeR to get differentially expressed genes (DEGs). With these DEGs, DEbPeak provides six differents charts to visualize these DEGs (VolcanoPlot, ScatterPlot, MAPlot, RankPlot, GenePlot and DEHeatmap).
Example data
We will used the results output by Quality Control vignette.
# library
suppressWarnings(suppressMessages(library(DESeq2)))
suppressWarnings(suppressMessages(library(edgeR)))
suppressWarnings(suppressMessages(library(DEbPeak)))
# load data
load(file = "/home/songyabing/R/learn/tmp/DEbPeak/example.RData")Differential Expression Analysis
Before visualizing the DEGs, you should get DEGs first. To increase flexibility, DEbPeak is compatible with both DESeq2 and edgeR results. We provides standard commands to get DEGs with both DESeq2 and edgeR.
DESeq2
# set control level
dds$condition <- relevel(dds$condition, ref = "WT")
# conduct differential expressed genes analysis
dds <- DESeq(dds)## estimating size factors## estimating dispersions## gene-wise dispersion estimates## mean-dispersion relationship## final dispersion estimates## fitting model and testing
# extract results
dds.results <- results(dds,contrast=c("condition",'KO','WT'))
dds.results.ordered <- dds.results[order(dds.results$log2FoldChange,decreasing = TRUE),]
head(dds.results.ordered)## log2 fold change (MLE): condition KO vs WT
## Wald test p-value: condition KO vs WT
## DataFrame with 6 rows and 6 columns
## baseMean log2FoldChange lfcSE stat pvalue
## <numeric> <numeric> <numeric> <numeric> <numeric>
## ENSMUSG00000025934.15 2.01093 4.46030 3.01742 1.47818 NA
## ENSMUSG00000105597.1 1.64376 4.16931 2.98872 1.39502 0.163011113
## ENSMUSG00000109866.1 2.32064 4.14573 1.22504 3.38415 0.000713987
## ENSMUSG00000085819.7 2.23441 4.07496 1.65939 2.45569 0.014061354
## ENSMUSG00000102474.1 1.52867 4.06781 1.80781 2.25013 0.024440396
## ENSMUSG00000042677.7 1.49781 4.05394 1.71350 2.36588 0.017987495
## padj
## <numeric>
## ENSMUSG00000025934.15 NA
## ENSMUSG00000105597.1 0.99973
## ENSMUSG00000109866.1 0.99973
## ENSMUSG00000085819.7 0.99973
## ENSMUSG00000102474.1 0.99973
## ENSMUSG00000042677.7 0.99973edgeR
snon.edgeR=DGEList(counts=count.matrix, group=meta.info$condition)
keep <- filterByExpr(snon.edgeR,min.count=10)
snon.edgeR <- snon.edgeR[keep, , keep.lib.sizes=FALSE]
snon.edgeR <- calcNormFactors(snon.edgeR)
snon.edgeR$samples$group <- relevel(snon.edgeR$samples$group, ref="WT")
design <- model.matrix(~snon.edgeR$samples$group)
snon.edgeR <- estimateDisp(snon.edgeR, design)
fit <- glmQLFit(snon.edgeR, design)
qlf <- glmQLFTest(fit, coef=2)
all.res <- topTags(qlf,n=nrow(snon.edgeR$counts))$table
head(all.res)## logFC logCPM F PValue FDR
## ENSMUSG00000039620.17 2.1925630 1.7076072 70.78781 2.686429e-07 0.003577786
## ENSMUSG00000046152.16 -0.7238661 4.9567684 36.89615 1.550592e-05 0.103253915
## ENSMUSG00000022376.8 -1.7356149 0.3243428 21.61086 2.617267e-04 0.998523400
## ENSMUSG00000027660.16 -0.9581197 5.3679317 19.98078 3.791045e-04 0.998523400
## ENSMUSG00000031478.16 0.8734604 2.4708238 18.99645 4.783255e-04 0.998523400
## ENSMUSG00000079469.10 -1.2129059 1.4296852 18.36878 5.568020e-04 0.998523400Visualization
VolcanoPlot
# VolcanoPlot for DESeq2
VolcanoPlot(dds.results.ordered,signif="pvalue",l2fc.threshold=0.3,label.num=2,
point.alpha = 0.8, label.color=c("purple","green"),tick.trans = NULL)## Differential expression analysis with DESeq2!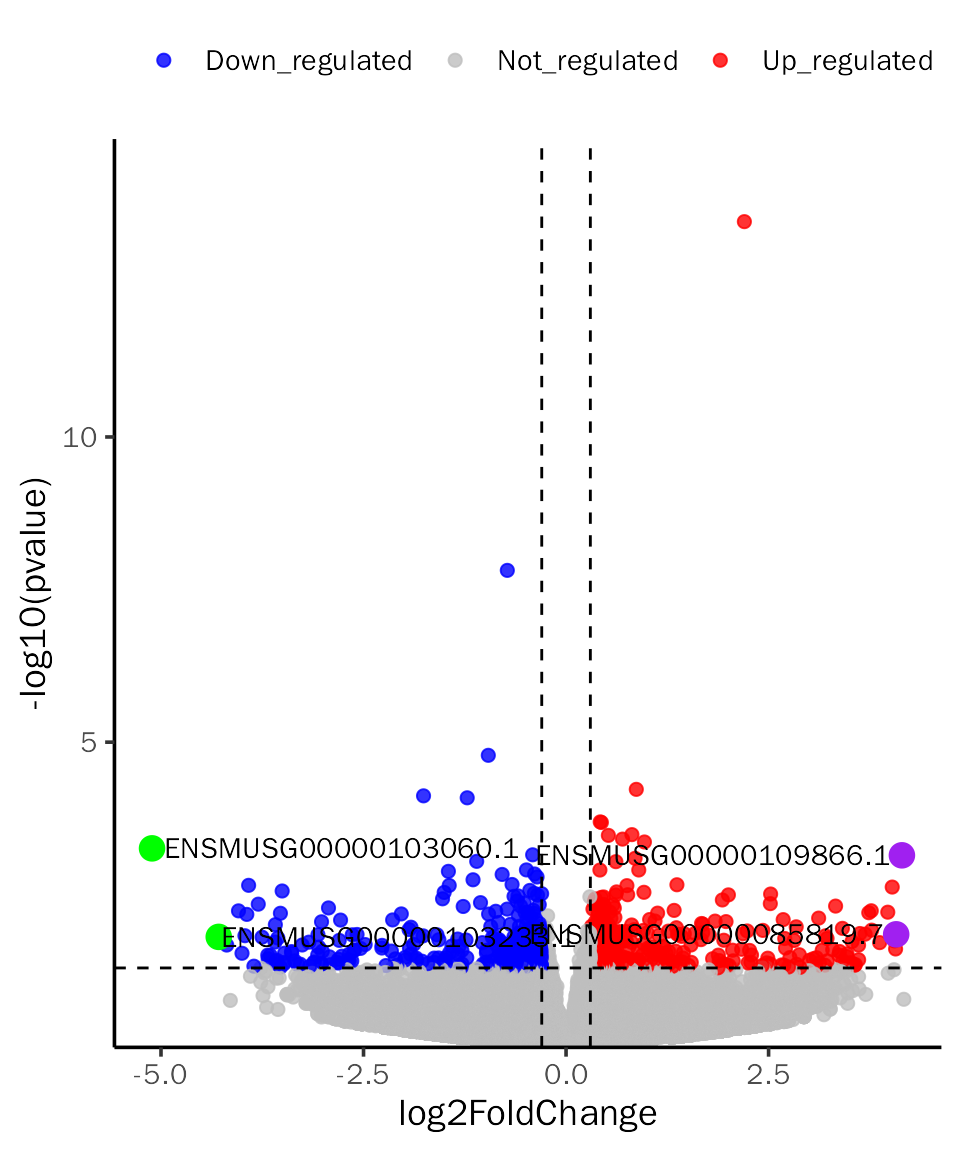
# VolcanoPlot for edgeR
VolcanoPlot(all.res,signif="PValue",l2fc.threshold=0.3,label.num=2,point.alpha = 0.8,
label.color=c("purple","green"),tick.trans = NULL)## Differential expression analysis with edgeR!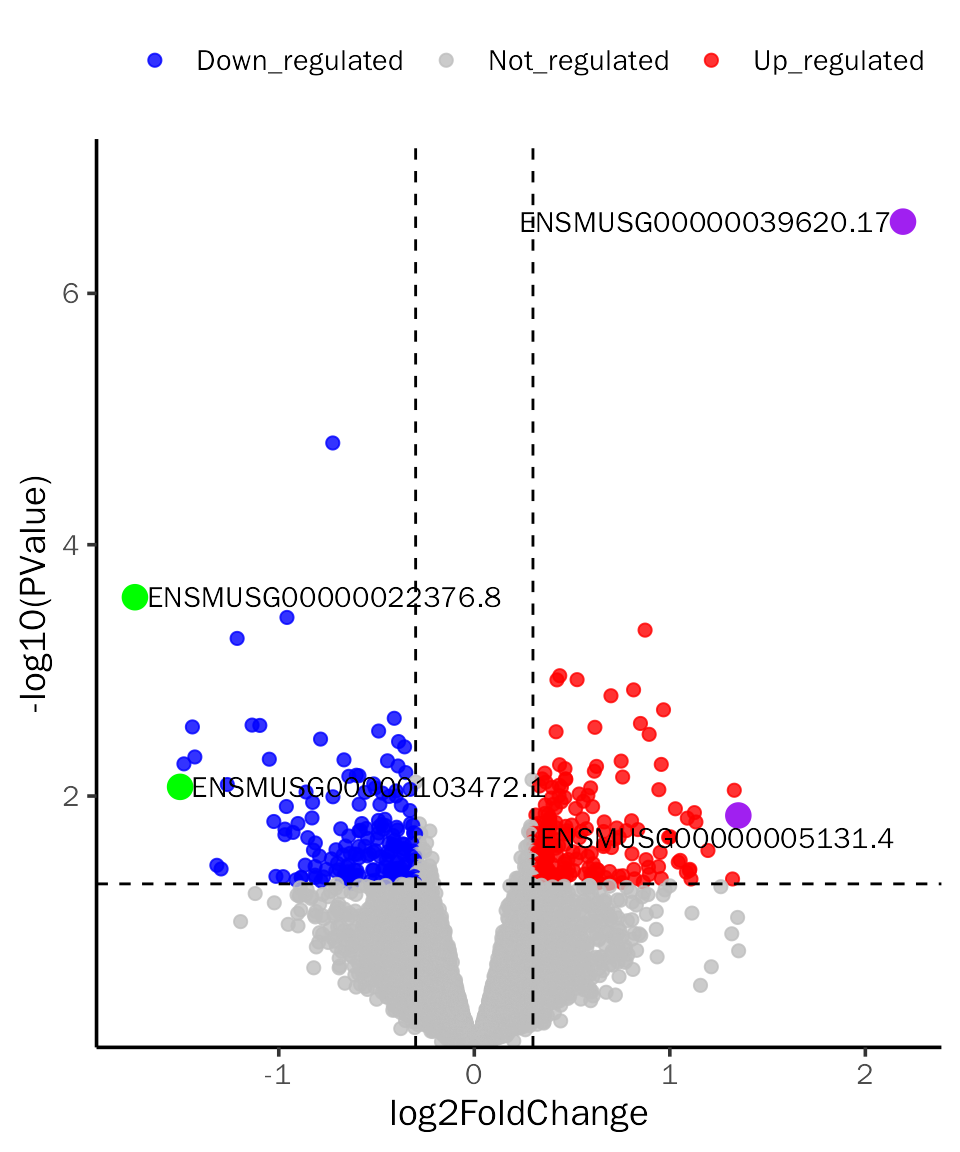
ScatterPlot
# ScatterPlot for DESeq2
ScatterPlot(deobj = dds,deres = dds.results.ordered,group.key = "condition",
ref.group = "WT",signif="pvalue",l2fc.threshold=0.3,label.num = 2,
point.alpha = 0.8,label.color=c("purple","green"))## Differential expression analysis with DESeq2!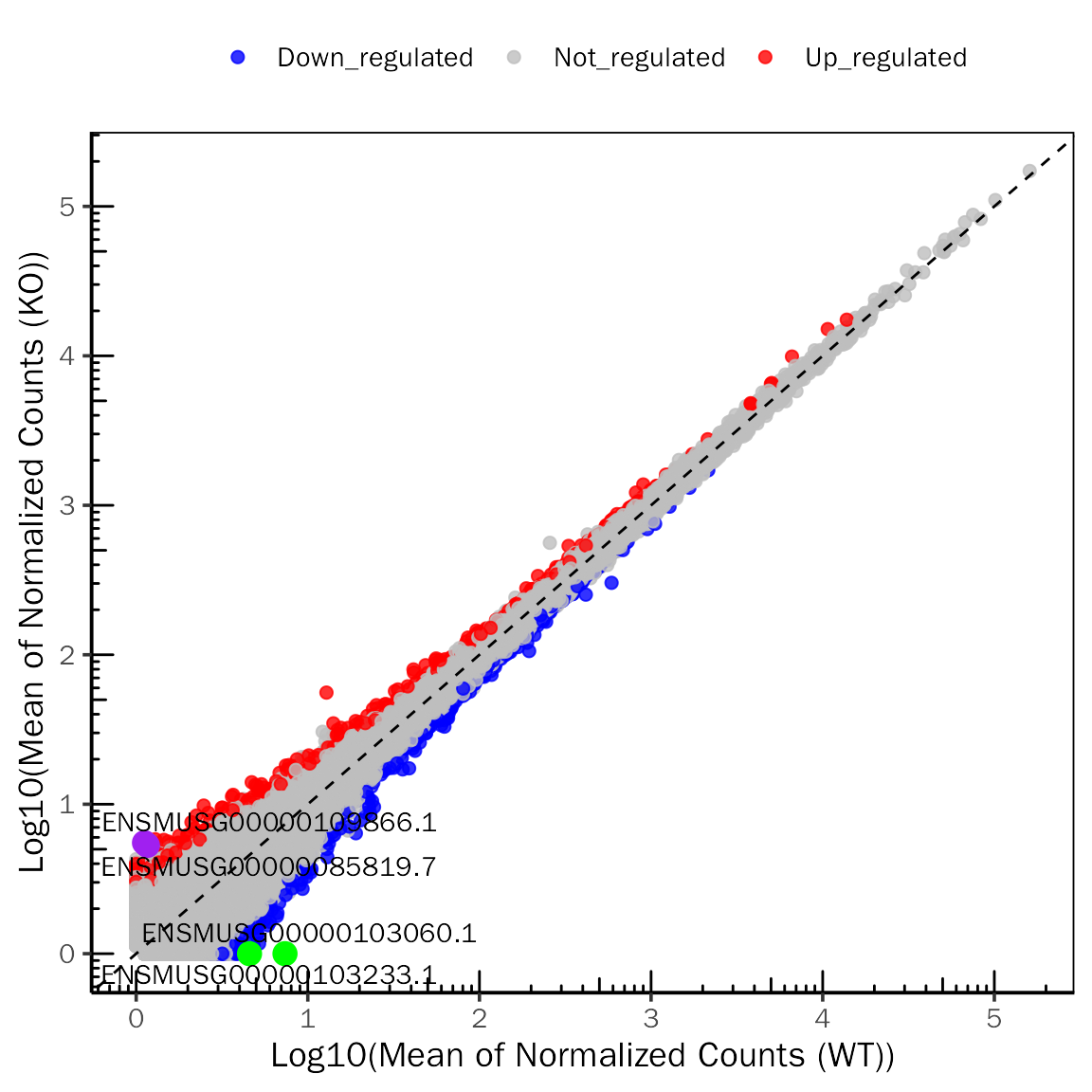
# ScatterPlot for edgeR
ScatterPlot(deobj = snon.edgeR,deres = all.res,group.key = "condition",
ref.group = "WT",signif="PValue",l2fc.threshold=0.3,label.num = 2,
point.alpha = 0.8,label.color=c("purple","green"))## Differential expression analysis with edgeR!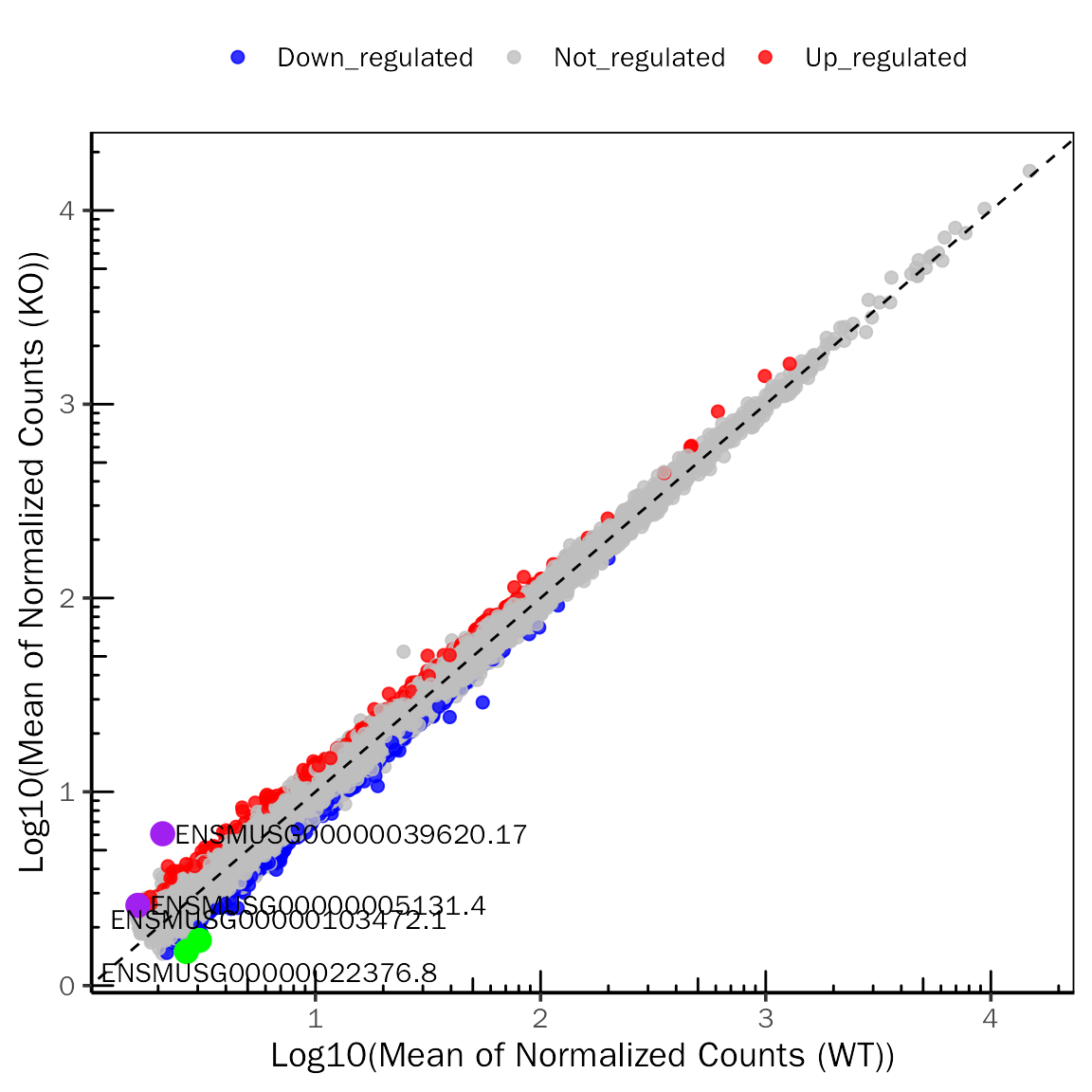
MAPlot
# MAPlot for DESeq2
MAPlot(dds.results.ordered,signif="pvalue",l2fc.threshold=0.3,label.num=2,
point.alpha = 0.8, label.color=c("purple","green"))## Differential expression analysis with DESeq2!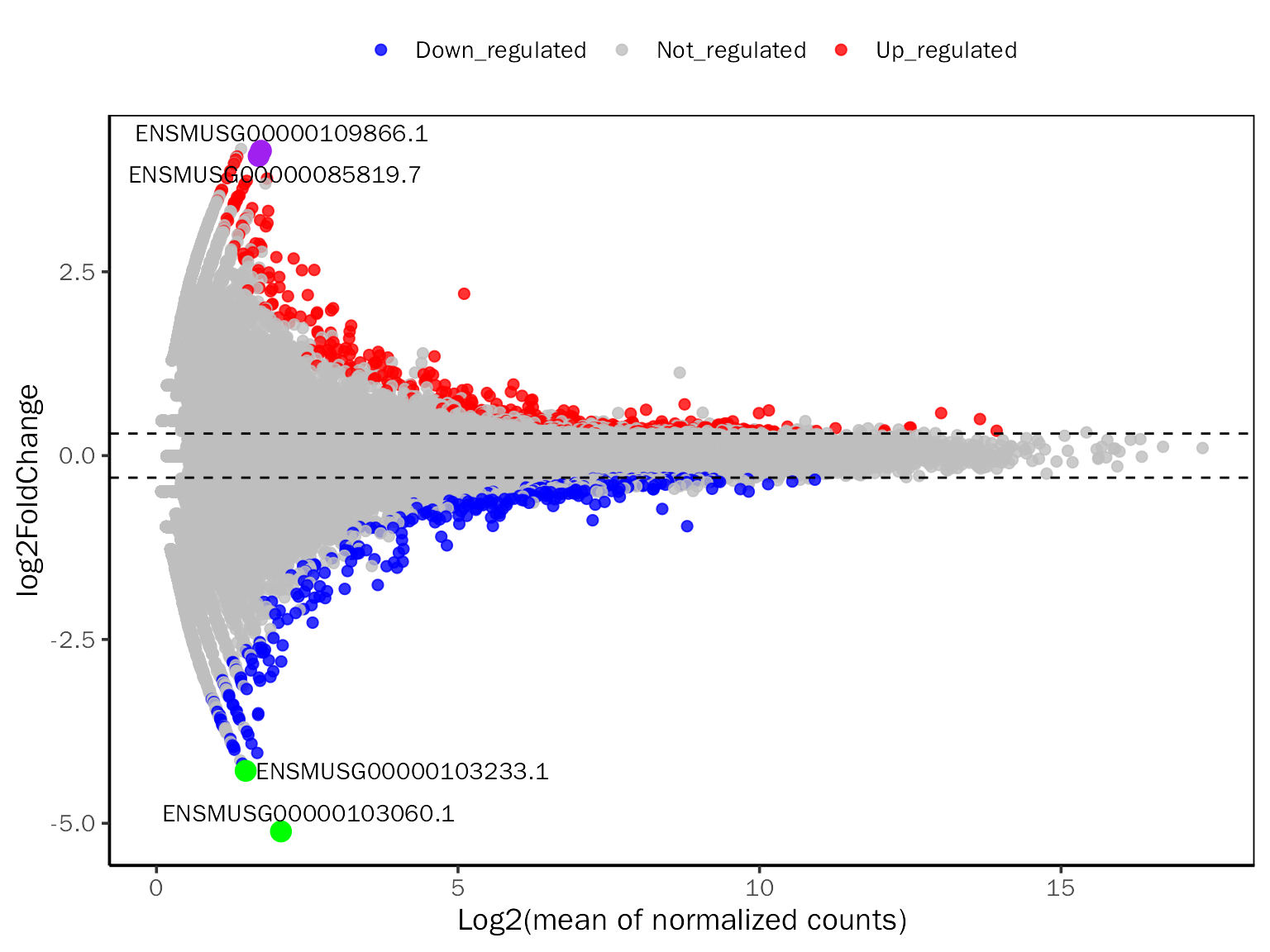
# MAPlot for edgeR
MAPlot(all.res,signif="PValue",l2fc.threshold=0.3,label.num=2,point.alpha = 0.8,
label.color=c("purple","green"))## Differential expression analysis with edgeR!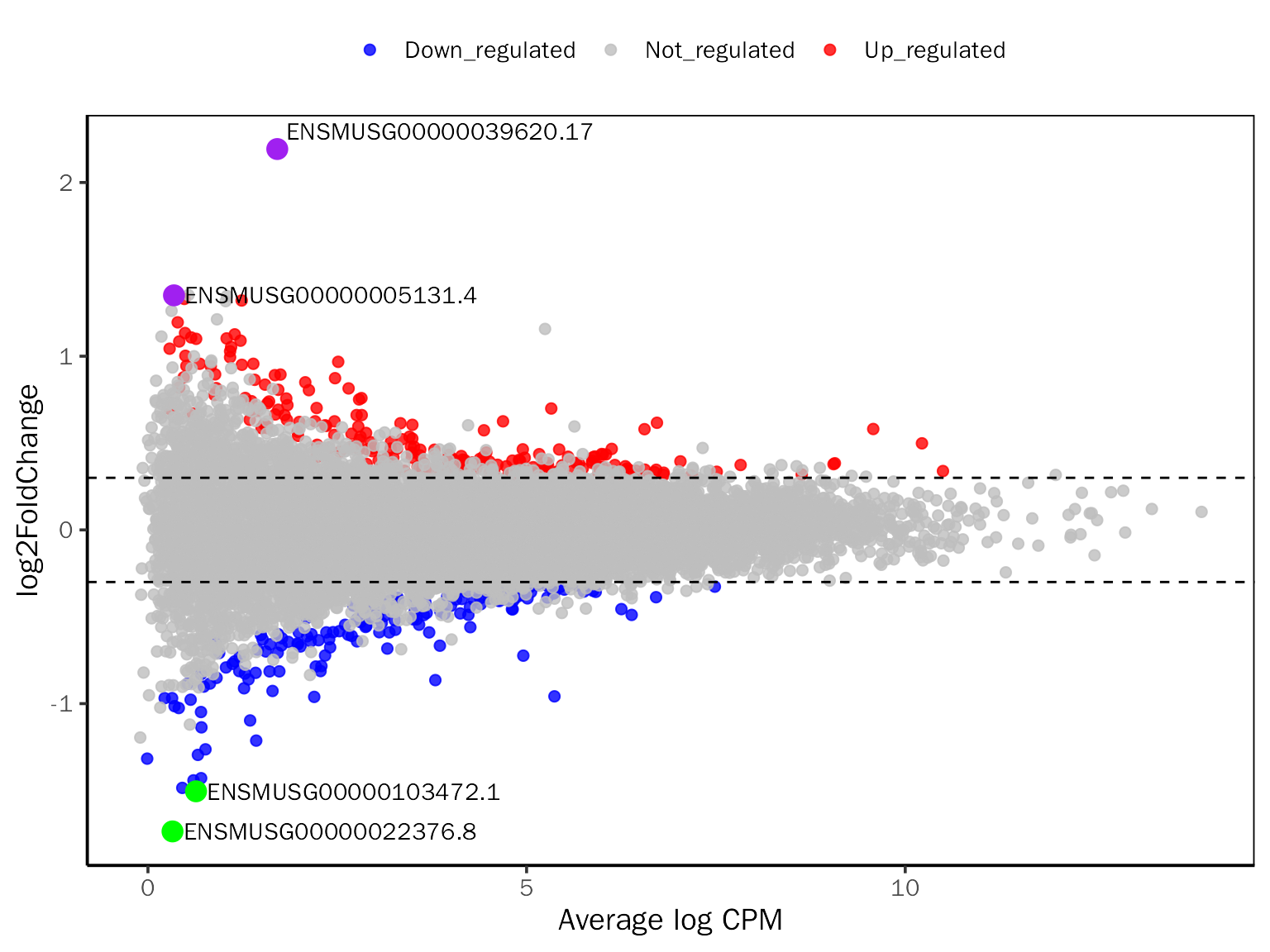
RankPlot
# RankPlot for DESeq2
RankPlot(dds.results.ordered,signif="pvalue",l2fc.threshold=0.3,label.num=2,
point.alpha = 0.8, label.color=c("purple","green"))## Differential expression analysis with DESeq2!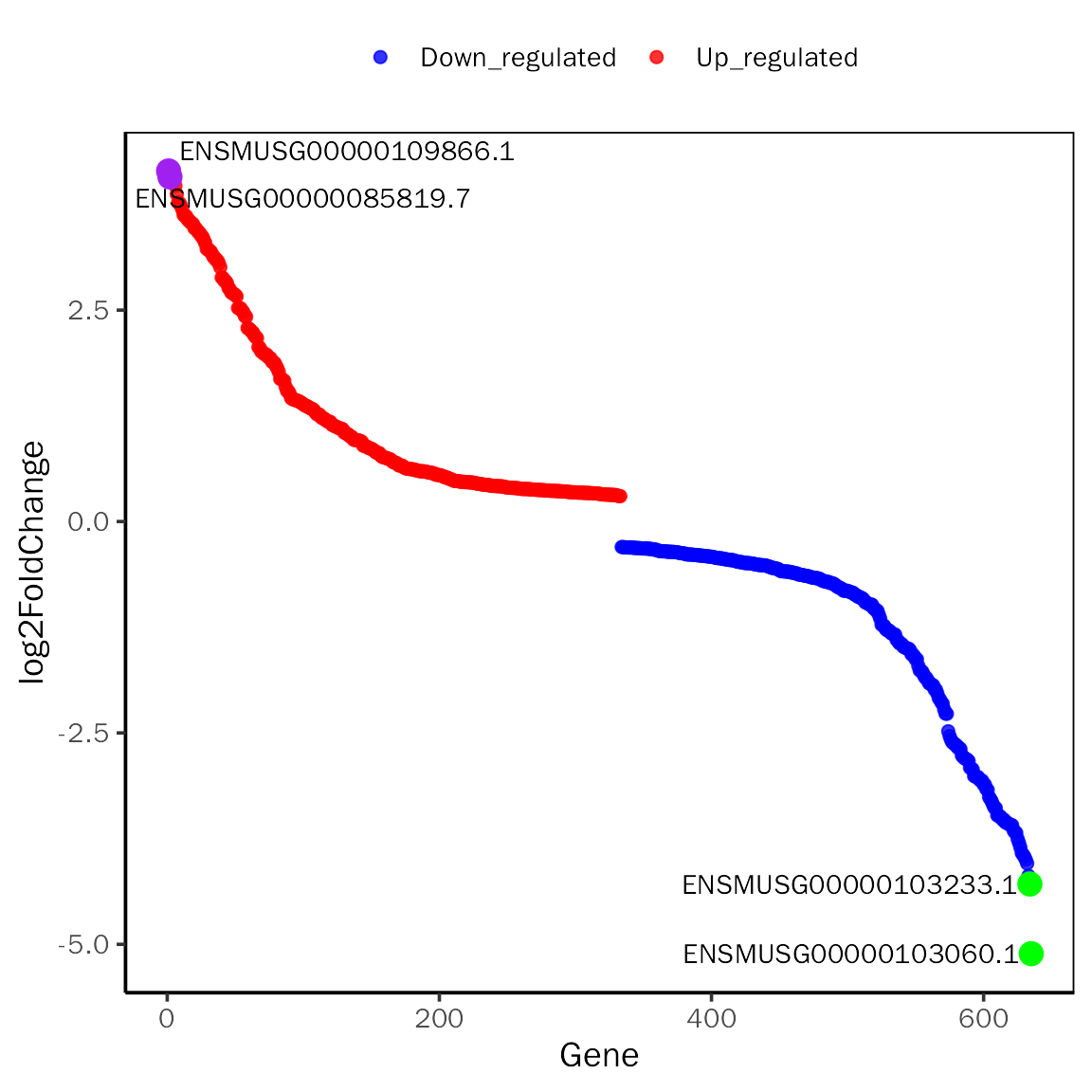
# RankPlot for edgeR
RankPlot(all.res,signif="PValue",l2fc.threshold=0.3,label.num=2,point.alpha = 0.8,
label.color=c("purple","green"))## Differential expression analysis with edgeR!
GenePlot
# GenePlot for DESeq2
GenePlot(deobj = dds,deres = dds.results.ordered,group.key = "condition",
ref.group = "WT",fill.color=c("red","blue"), fill.alpha = 0.8,
gene.num =2,signif="pvalue",l2fc.threshold=0.3)## Differential expression analysis with DESeq2!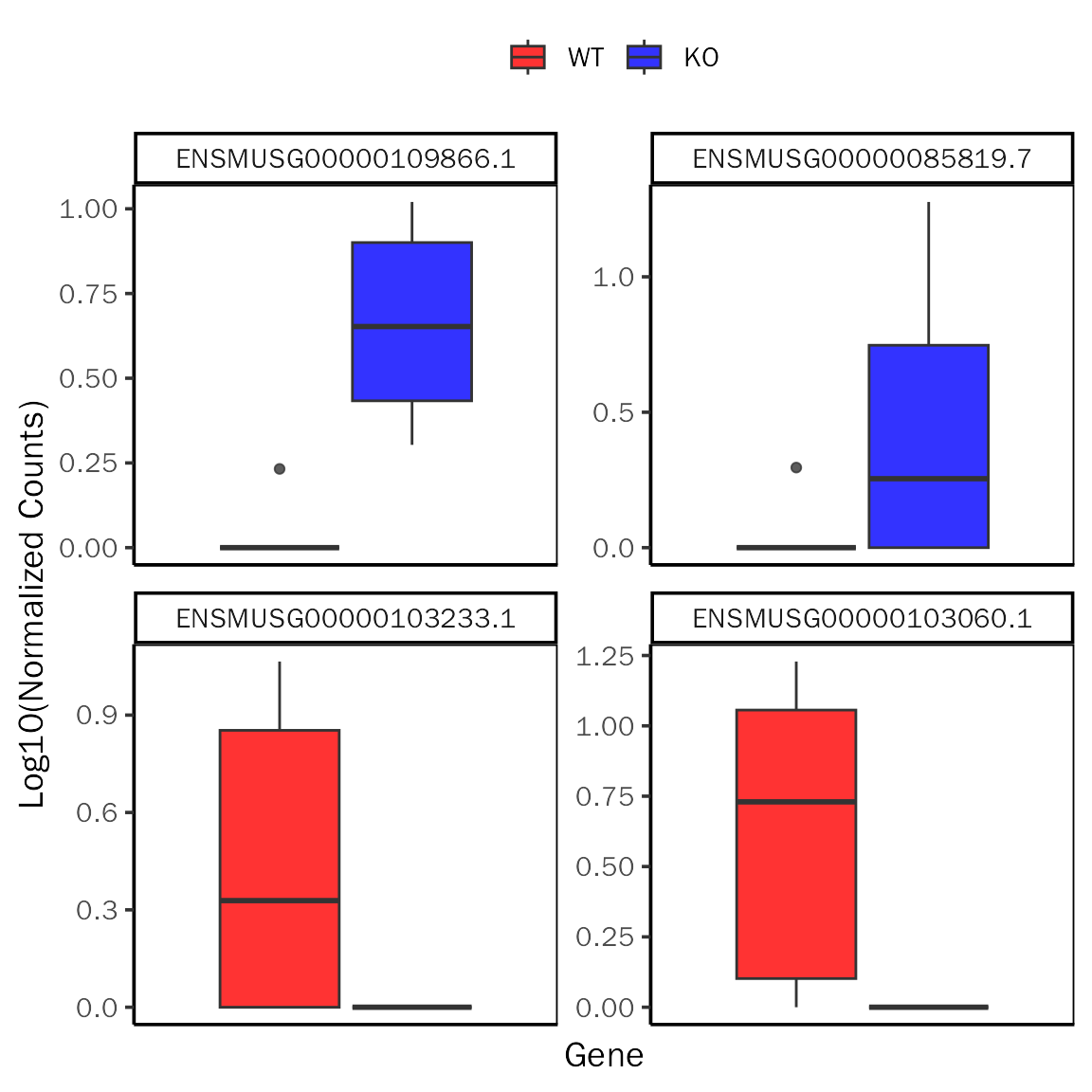
# GenePlot for edgeR
GenePlot(deobj = snon.edgeR,deres = all.res,group.key = "condition",
ref.group = "WT",fill.color=c("red","blue"),fill.alpha = 0.8,
gene.num =2,signif="PValue",l2fc.threshold=0.3)## Differential expression analysis with edgeR!
DEHeatmap
# DEHeatmap for DESeq2
DEHeatmap(deobj = dds,deres = dds.results.ordered,group.key = "condition",
ref.group = "WT", signif="pvalue",l2fc.threshold=0.3)## Differential expression analysis with DESeq2!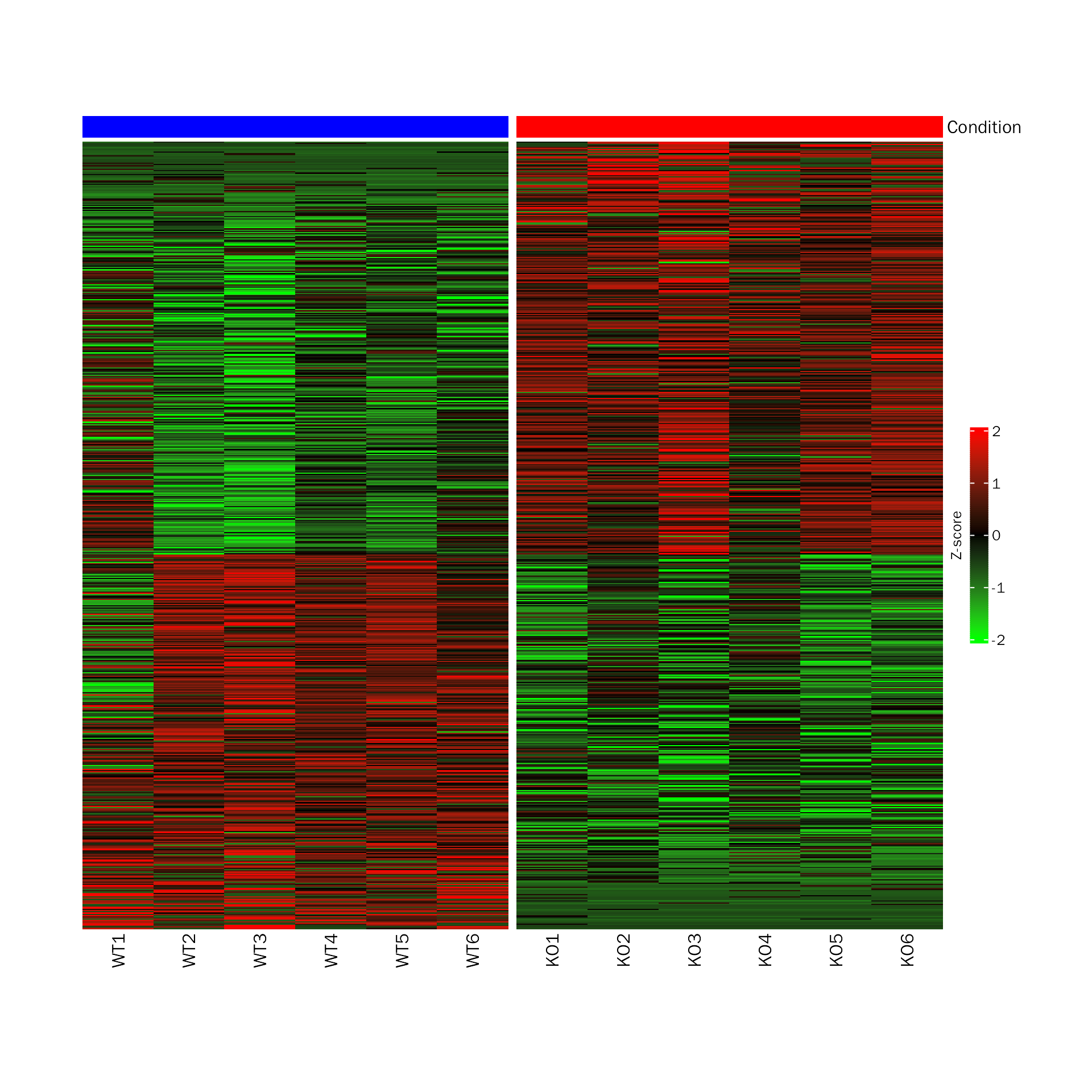
# DEHeatmap for edgeR
DEHeatmap(deobj = snon.edgeR,deres = all.res,group.key = "condition",
ref.group = "WT", signif="PValue",l2fc.threshold=0.3)## Differential expression analysis with edgeR!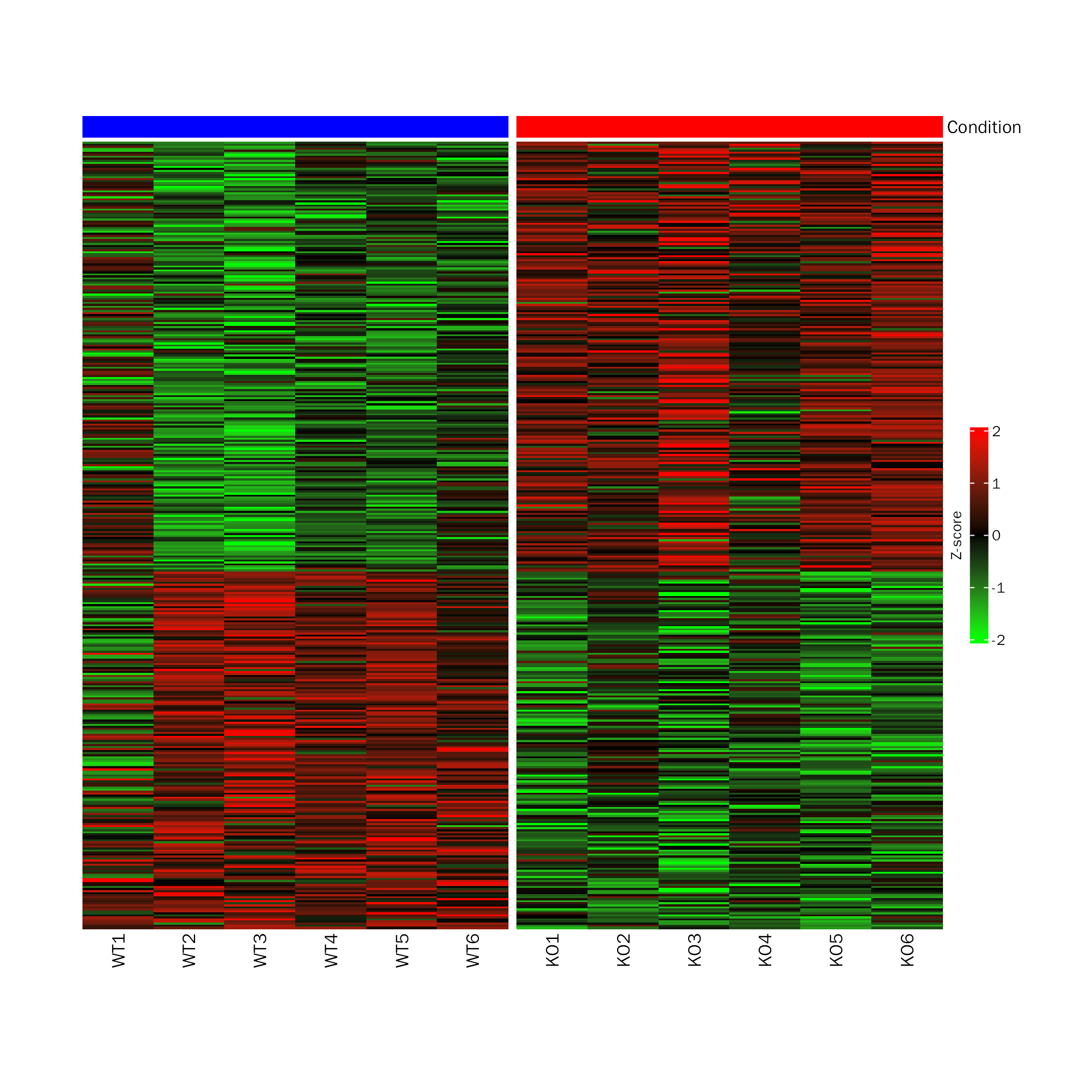
Session info
## R version 4.0.3 (2020-10-10)
## Platform: x86_64-conda-linux-gnu (64-bit)
## Running under: CentOS Linux 7 (Core)
##
## Matrix products: default
## BLAS/LAPACK: /home/softwares/anaconda3/envs/r4.0/lib/libopenblasp-r0.3.12.so
##
## locale:
## [1] LC_CTYPE=zh_CN.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=zh_CN.UTF-8 LC_COLLATE=zh_CN.UTF-8
## [5] LC_MONETARY=zh_CN.UTF-8 LC_MESSAGES=zh_CN.UTF-8
## [7] LC_PAPER=zh_CN.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=zh_CN.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] DEbPeak_1.4.0 edgeR_3.32.1
## [3] limma_3.46.0 DESeq2_1.30.1
## [5] SummarizedExperiment_1.20.0 Biobase_2.50.0
## [7] MatrixGenerics_1.2.1 matrixStats_0.58.0
## [9] GenomicRanges_1.42.0 GenomeInfoDb_1.26.7
## [11] IRanges_2.24.1 S4Vectors_0.28.1
## [13] BiocGenerics_0.42.0
##
## loaded via a namespace (and not attached):
## [1] rsvd_1.0.3
## [2] ggvenn_0.1.9
## [3] apeglm_1.12.0
## [4] Rsamtools_2.6.0
## [5] rsvg_2.1
## [6] foreach_1.5.1
## [7] rprojroot_2.0.2
## [8] crayon_1.4.1
## [9] V8_3.4.2
## [10] MASS_7.3-58
## [11] nlme_3.1-152
## [12] backports_1.2.1
## [13] sva_3.38.0
## [14] GOSemSim_2.25.0
## [15] rlang_1.1.0
## [16] XVector_0.30.0
## [17] readxl_1.4.2
## [18] irlba_2.3.5
## [19] GOstats_2.56.0
## [20] BiocParallel_1.24.1
## [21] rjson_0.2.20
## [22] bit64_4.0.5
## [23] glue_1.6.2
## [24] DiffBind_3.0.15
## [25] mixsqp_0.3-43
## [26] pheatmap_1.0.12
## [27] parallel_4.0.3
## [28] AnnotationDbi_1.52.0
## [29] DEFormats_1.18.0
## [30] base64url_1.4
## [31] tcltk_4.0.3
## [32] DOSE_3.23.2
## [33] haven_2.5.2
## [34] tidyselect_1.2.0
## [35] rio_0.5.27
## [36] XML_3.99-0.6
## [37] tidyr_1.3.0
## [38] ggpubr_0.4.0
## [39] GenomicAlignments_1.26.0
## [40] xtable_1.8-4
## [41] ggnetwork_0.5.12
## [42] magrittr_2.0.3
## [43] evaluate_0.14
## [44] ggplot2_3.4.2
## [45] cli_3.6.1
## [46] zlibbioc_1.36.0
## [47] hwriter_1.3.2
## [48] rstudioapi_0.14
## [49] bslib_0.3.1
## [50] GreyListChIP_1.22.0
## [51] fastmatch_1.1-3
## [52] BiocSingular_1.6.0
## [53] xfun_0.30
## [54] askpass_1.1
## [55] clue_0.3-59
## [56] gson_0.0.9
## [57] cluster_2.1.1
## [58] caTools_1.18.2
## [59] tidygraph_1.2.0
## [60] tibble_3.2.1
## [61] ggrepel_0.9.1
## [62] Biostrings_2.58.0
## [63] png_0.1-7
## [64] withr_2.5.0
## [65] bitops_1.0-6
## [66] ggforce_0.3.3
## [67] RBGL_1.66.0
## [68] plyr_1.8.6
## [69] cellranger_1.1.0
## [70] GSEABase_1.52.1
## [71] pcaPP_2.0-1
## [72] dqrng_0.2.1
## [73] coda_0.19-4
## [74] pillar_1.9.0
## [75] gplots_3.1.1
## [76] GlobalOptions_0.1.2
## [77] cachem_1.0.4
## [78] GenomicFeatures_1.42.2
## [79] fs_1.5.0
## [80] GetoptLong_1.0.5
## [81] clusterProfiler_4.7.1
## [82] DelayedMatrixStats_1.12.3
## [83] vctrs_0.6.2
## [84] generics_0.1.0
## [85] plot3D_1.4
## [86] tools_4.0.3
## [87] foreign_0.8-81
## [88] NOISeq_2.34.0
## [89] munsell_0.5.0
## [90] tweenr_1.0.2
## [91] fgsea_1.16.0
## [92] DelayedArray_0.16.3
## [93] fastmap_1.1.0
## [94] compiler_4.0.3
## [95] abind_1.4-5
## [96] rtracklayer_1.50.0
## [97] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
## [98] GenomeInfoDbData_1.2.4
## [99] gridExtra_2.3
## [100] lattice_0.20-45
## [101] ggnewscale_0.4.7
## [102] AnnotationForge_1.32.0
## [103] utf8_1.2.1
## [104] dplyr_1.1.2
## [105] BiocFileCache_1.14.0
## [106] jsonlite_1.8.4
## [107] scales_1.2.1
## [108] graph_1.68.0
## [109] carData_3.0-4
## [110] sparseMatrixStats_1.2.1
## [111] TFEA.ChIP_1.10.0
## [112] genefilter_1.72.1
## [113] car_3.0-11
## [114] doParallel_1.0.16
## [115] latticeExtra_0.6-29
## [116] R.utils_2.12.0
## [117] brew_1.0-6
## [118] checkmate_2.0.0
## [119] rmarkdown_2.14
## [120] openxlsx_4.2.3
## [121] pkgdown_1.6.1
## [122] cowplot_1.1.1
## [123] textshaping_0.3.6
## [124] forcats_1.0.0
## [125] downloader_0.4
## [126] BSgenome_1.58.0
## [127] igraph_1.4.99.9024
## [128] survival_3.2-10
## [129] numDeriv_2016.8-1.1
## [130] yaml_2.2.1
## [131] plotrix_3.8-2
## [132] systemfonts_1.0.4
## [133] ashr_2.2-47
## [134] SQUAREM_2021.1
## [135] htmltools_0.5.2
## [136] memoise_2.0.0
## [137] VariantAnnotation_1.36.0
## [138] locfit_1.5-9.4
## [139] graphlayouts_0.7.1
## [140] batchtools_0.9.15
## [141] PCAtools_2.2.0
## [142] viridisLite_0.4.0
## [143] rrcov_1.7-0
## [144] digest_0.6.27
## [145] assertthat_0.2.1
## [146] rappdirs_0.3.3
## [147] emdbook_1.3.12
## [148] RSQLite_2.2.5
## [149] amap_0.8-18
## [150] yulab.utils_0.0.4
## [151] debugme_1.1.0
## [152] misc3d_0.9-1
## [153] data.table_1.14.2
## [154] blob_1.2.1
## [155] R.oo_1.24.0
## [156] ragg_0.4.0
## [157] labeling_0.4.2
## [158] splines_4.0.3
## [159] Cairo_1.5-12.2
## [160] ggupset_0.3.0
## [161] RCurl_1.98-1.3
## [162] broom_1.0.4
## [163] hms_1.1.3
## [164] colorspace_2.0-0
## [165] BiocManager_1.30.16
## [166] shape_1.4.6
## [167] sass_0.4.1
## [168] GEOquery_2.58.0
## [169] Rcpp_1.0.9
## [170] mvtnorm_1.1-2
## [171] circlize_0.4.15
## [172] enrichplot_1.10.2
## [173] fansi_0.4.2
## [174] tzdb_0.3.0
## [175] truncnorm_1.0-8
## [176] ChIPseeker_1.33.0.900
## [177] R6_2.5.0
## [178] grid_4.0.3
## [179] lifecycle_1.0.3
## [180] ShortRead_1.48.0
## [181] zip_2.1.1
## [182] curl_4.3
## [183] ggsignif_0.6.3
## [184] jquerylib_0.1.3
## [185] robustbase_0.95-0
## [186] DO.db_2.9
## [187] Matrix_1.5-4
## [188] qvalue_2.22.0
## [189] desc_1.3.0
## [190] org.Hs.eg.db_3.12.0
## [191] RColorBrewer_1.1-2
## [192] iterators_1.0.13
## [193] stringr_1.5.0
## [194] DOT_0.1
## [195] ggpie_0.2.5
## [196] beachmat_2.6.4
## [197] polyclip_1.10-0
## [198] biomaRt_2.46.3
## [199] purrr_1.0.1
## [200] shadowtext_0.0.9
## [201] gridGraphics_0.5-1
## [202] mgcv_1.8-34
## [203] ComplexHeatmap_2.13.1
## [204] openssl_1.4.3
## [205] patchwork_1.0.0
## [206] bdsmatrix_1.3-4
## [207] codetools_0.2-18
## [208] invgamma_1.1
## [209] GO.db_3.12.1
## [210] gtools_3.8.2
## [211] prettyunits_1.1.1
## [212] dbplyr_2.3.2
## [213] R.methodsS3_1.8.1
## [214] gtable_0.3.0
## [215] DBI_1.1.1
## [216] highr_0.8
## [217] ggfun_0.0.6
## [218] httr_1.4.5
## [219] KernSmooth_2.23-18
## [220] stringi_1.5.3
## [221] progress_1.2.2
## [222] reshape2_1.4.4
## [223] farver_2.1.0
## [224] annotate_1.68.0
## [225] viridis_0.6.1
## [226] Rgraphviz_2.34.0
## [227] xml2_1.3.4
## [228] bbmle_1.0.24
## [229] systemPipeR_1.24.3
## [230] boot_1.3-28
## [231] readr_2.1.4
## [232] geneplotter_1.68.0
## [233] ggplotify_0.1.0
## [234] Category_2.56.0
## [235] DEoptimR_1.0-11
## [236] bit_4.0.4
## [237] scatterpie_0.1.7
## [238] jpeg_0.1-8.1
## [239] ggraph_2.0.5
## [240] pkgconfig_2.0.3
## [241] rstatix_0.7.0
## [242] knitr_1.37