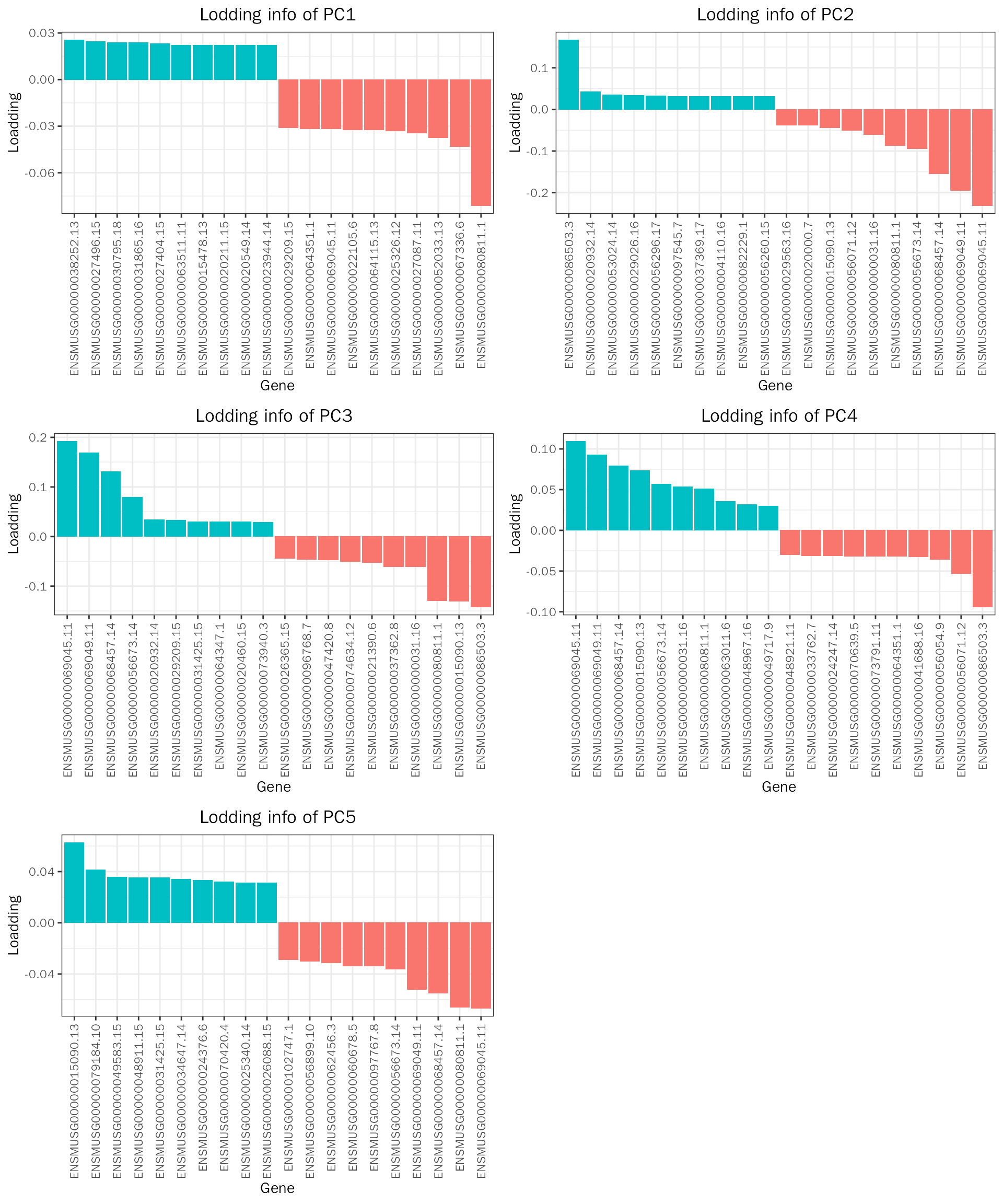
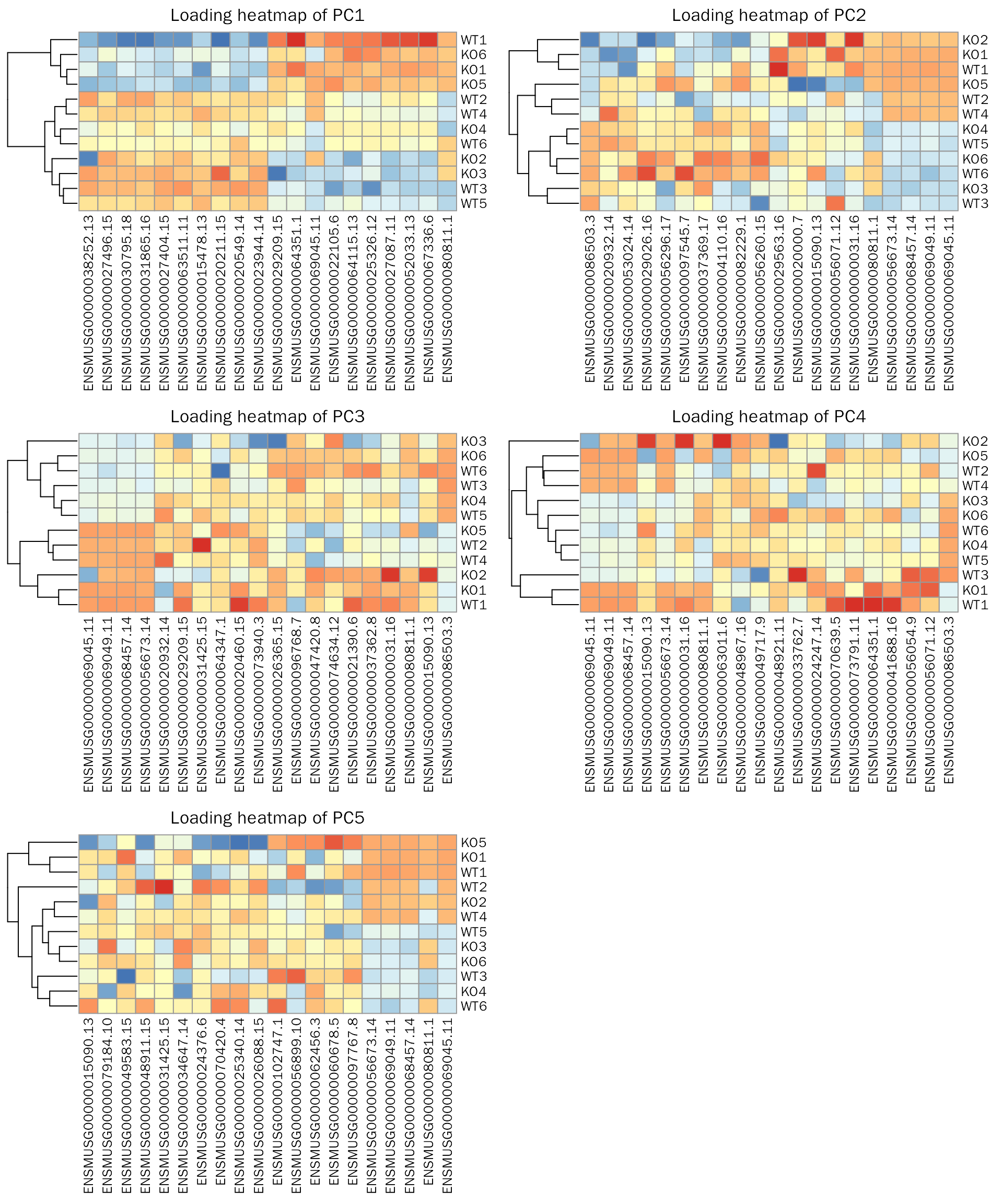
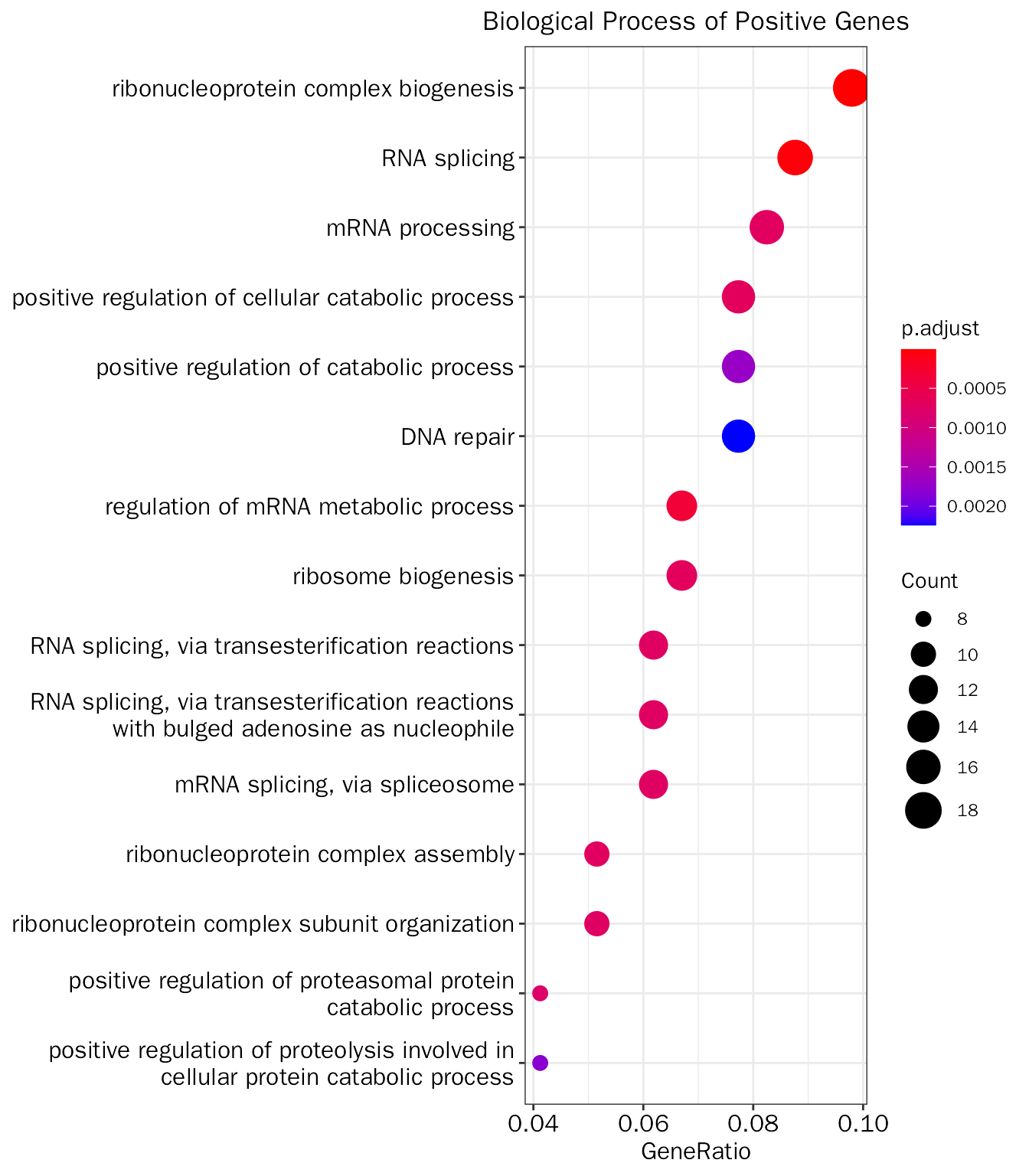
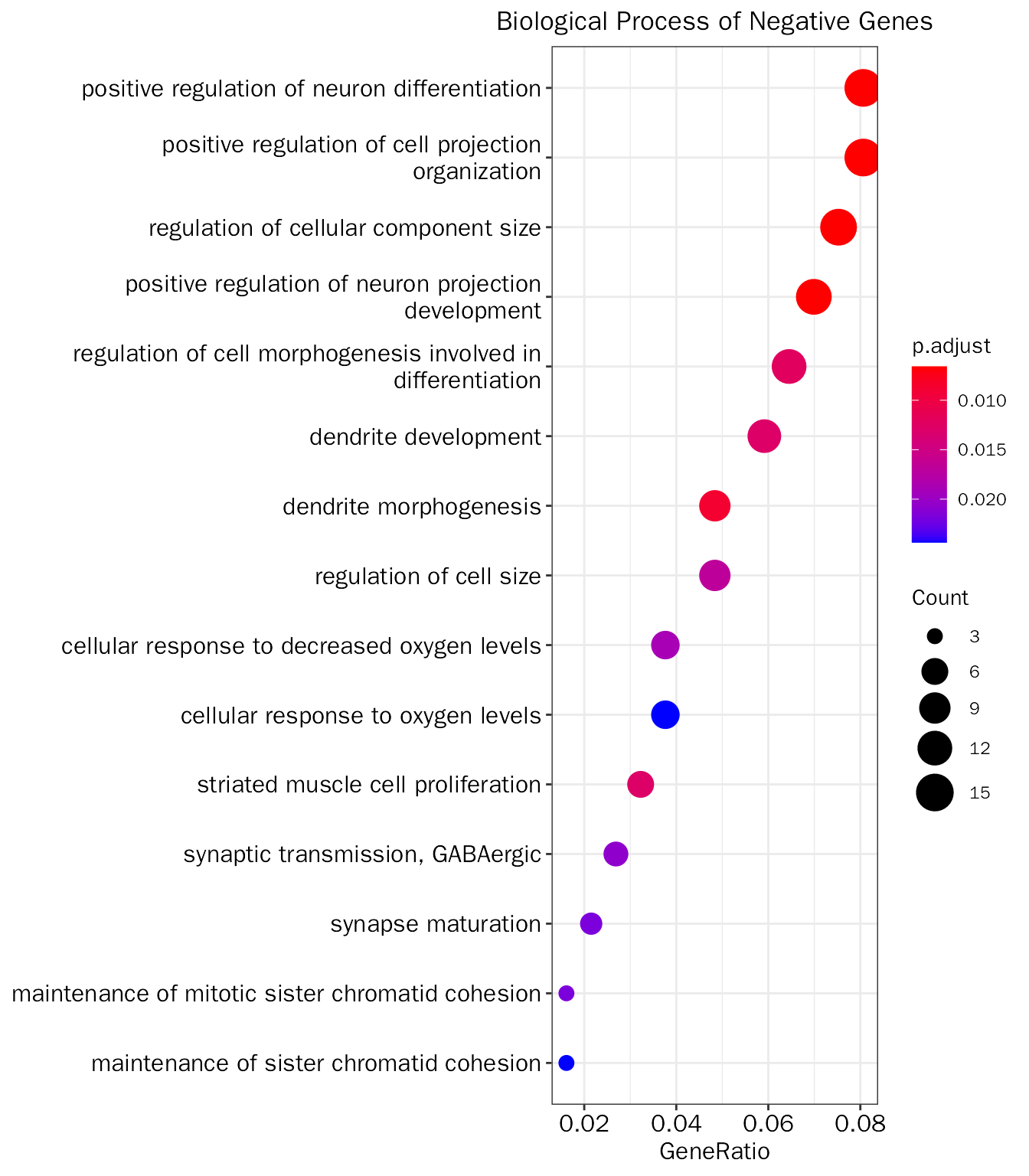
PrincipalComponentAnalysis
PrincipalComponentAnalysis.RmdIntroduction
Principal component analysis (PCA) is an unsupervised linear dimensionality reduction algorithm, and it can retain most of the variation in the data set. Based on these properties, PCA has two main uses in RNA-seq analysis:
- Quality control:
- Evaluate sample similarity across different conditions
- Check batch information
- Detect outliers
- Reveal biologically relevant information
For Quality Control, We have already covered in the previous vignette, here, we will focus on the “Reveal biologically relevant information”!
Example data
This vignette follows the previous “Quality Control” vignette, so we will used the results output by “Quality Control” vignette.
# library
suppressWarnings(suppressMessages(library(DEbPeak)))
# load data
load(file = "/home/songyabing/R/learn/tmp/DEbPeak/example.RData")PCA
We will perform PCA with prcomp, and remove outlier samples detected by previous QCPCA.
# conduct PCA
pca.res=PCA(deobj = dds,remove.sample = outlier.res$outlier,transform.method = "rlog")
# get basic plots
basic.plots=PCABasic(pca.res,colby="condition",legend.pos = "right")Scree plot
A Scree Plot is a simple line plot that shows the total amount of variance that can be explained by each individual PC (Y-axis shows explained variance, X-axis shows the number of PCs). It can be used to determine the number of PCs to be explored for downstream analysis. Here, we created a cumulative scree plot based on PCAtools, the red dashed line represents 90% explained variance.
basic.plots[["screen"]]
Biplot
Biplot contains informations from two aspects:
- sample similarity (point)
- how strongly each gene influences a principal component (vector)
- the vector will have projections on PCs, and the project values on each PC show how much weight they have on that PC. Positive values indicate that a vector/gene and a principal component are positively correlated whereas negative values indicate a negative correlation.
- the angle between vectors indicates how vectors/genes correlate with one another: a small angle implies positive correlation, a large one suggests negative correlation, and a 90 degree angle indicates no correlation between two vectors/genes.
The biplot is created based on PCAtools:
basic.plots[["biplot"]]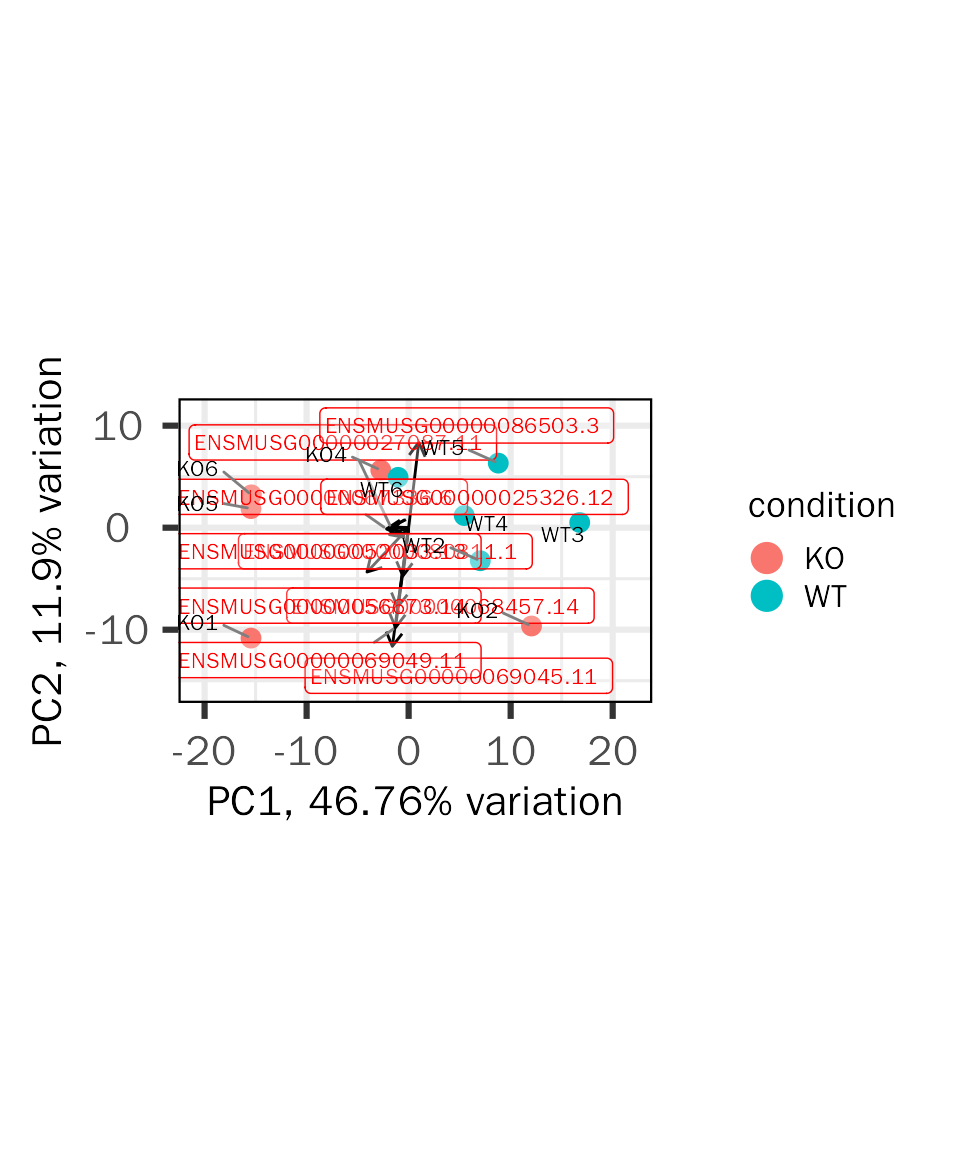
PC pairs plot
The PC pairs plot (based on PCAtools) will show sample similarity across different PC combination:
basic.plots[["pairs"]]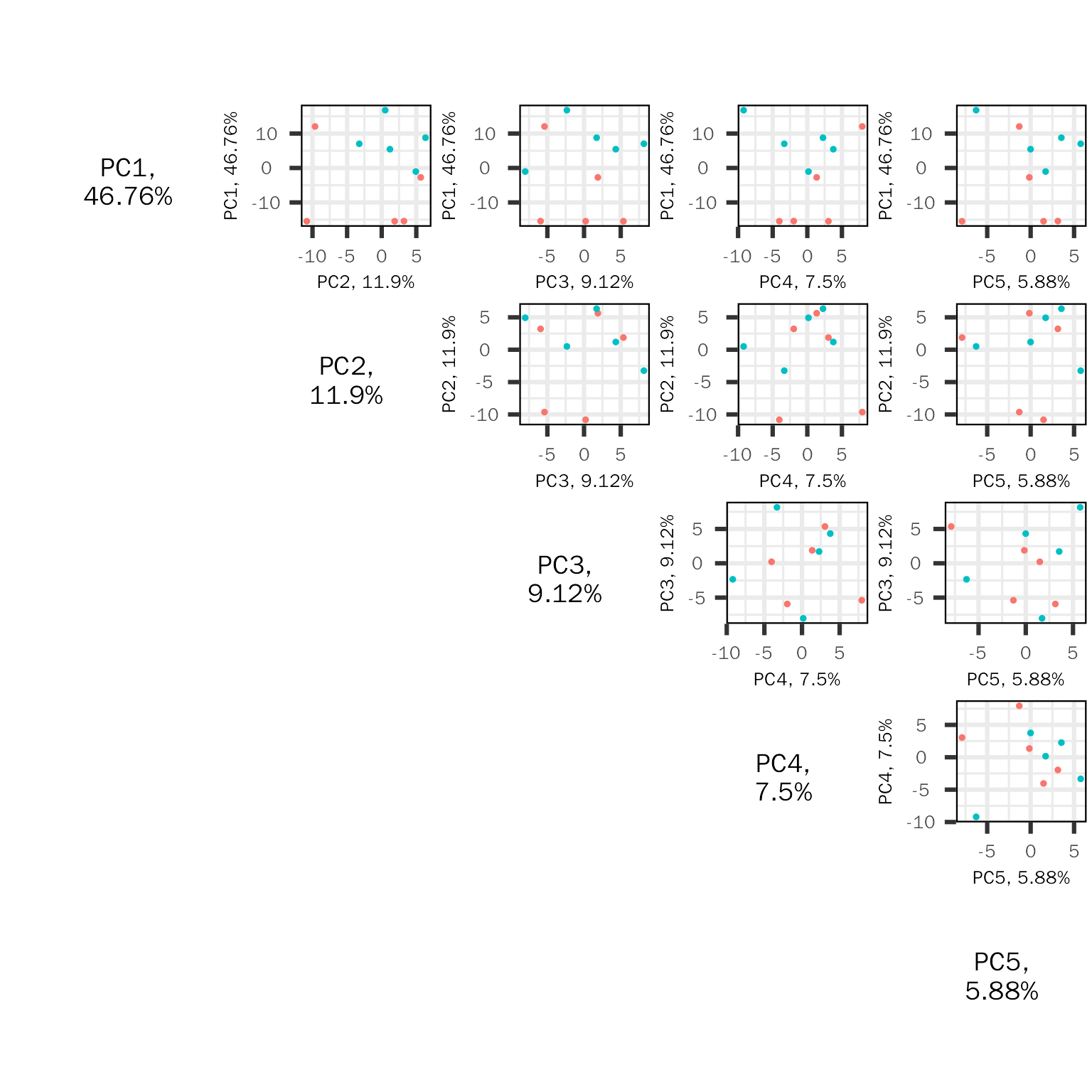
3D visualization
To visualize three PCs simultaneously, DEbPeak provides PCA3D to create 3D PCA plot:
PCA3D(pca = pca.res,color.key = "condition",main = "3D PCA")
Session info
## R version 4.0.3 (2020-10-10)
## Platform: x86_64-conda-linux-gnu (64-bit)
## Running under: CentOS Linux 7 (Core)
##
## Matrix products: default
## BLAS/LAPACK: /home/softwares/anaconda3/envs/r4.0/lib/libopenblasp-r0.3.12.so
##
## locale:
## [1] LC_CTYPE=zh_CN.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=zh_CN.UTF-8 LC_COLLATE=zh_CN.UTF-8
## [5] LC_MONETARY=zh_CN.UTF-8 LC_MESSAGES=zh_CN.UTF-8
## [7] LC_PAPER=zh_CN.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=zh_CN.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] org.Mm.eg.db_3.12.0 AnnotationDbi_1.52.0 IRanges_2.24.1
## [4] S4Vectors_0.28.1 Biobase_2.50.0 BiocGenerics_0.42.0
## [7] DEbPeak_1.4.0
##
## loaded via a namespace (and not attached):
## [1] rsvd_1.0.3
## [2] ggvenn_0.1.9
## [3] apeglm_1.12.0
## [4] Rsamtools_2.6.0
## [5] rsvg_2.1
## [6] foreach_1.5.1
## [7] rprojroot_2.0.2
## [8] crayon_1.4.1
## [9] V8_3.4.2
## [10] MASS_7.3-58
## [11] nlme_3.1-152
## [12] backports_1.2.1
## [13] sva_3.38.0
## [14] GOSemSim_2.25.0
## [15] rlang_1.1.0
## [16] XVector_0.30.0
## [17] readxl_1.4.2
## [18] irlba_2.3.5
## [19] limma_3.46.0
## [20] GOstats_2.56.0
## [21] BiocParallel_1.24.1
## [22] rjson_0.2.20
## [23] bit64_4.0.5
## [24] glue_1.6.2
## [25] DiffBind_3.0.15
## [26] mixsqp_0.3-43
## [27] pheatmap_1.0.12
## [28] parallel_4.0.3
## [29] DEFormats_1.18.0
## [30] base64url_1.4
## [31] tcltk_4.0.3
## [32] DOSE_3.23.2
## [33] haven_2.5.2
## [34] tidyselect_1.2.0
## [35] SummarizedExperiment_1.20.0
## [36] rio_0.5.27
## [37] XML_3.99-0.6
## [38] tidyr_1.3.0
## [39] ggpubr_0.4.0
## [40] GenomicAlignments_1.26.0
## [41] xtable_1.8-4
## [42] ggnetwork_0.5.12
## [43] magrittr_2.0.3
## [44] evaluate_0.14
## [45] ggplot2_3.4.2
## [46] cli_3.6.1
## [47] zlibbioc_1.36.0
## [48] hwriter_1.3.2
## [49] rstudioapi_0.14
## [50] bslib_0.3.1
## [51] GreyListChIP_1.22.0
## [52] fastmatch_1.1-3
## [53] BiocSingular_1.6.0
## [54] xfun_0.30
## [55] askpass_1.1
## [56] clue_0.3-59
## [57] gson_0.0.9
## [58] cluster_2.1.1
## [59] caTools_1.18.2
## [60] tidygraph_1.2.0
## [61] tibble_3.2.1
## [62] ggrepel_0.9.1
## [63] Biostrings_2.58.0
## [64] png_0.1-7
## [65] withr_2.5.0
## [66] bitops_1.0-6
## [67] ggforce_0.3.3
## [68] RBGL_1.66.0
## [69] plyr_1.8.6
## [70] cellranger_1.1.0
## [71] GSEABase_1.52.1
## [72] pcaPP_2.0-1
## [73] dqrng_0.2.1
## [74] coda_0.19-4
## [75] pillar_1.9.0
## [76] gplots_3.1.1
## [77] GlobalOptions_0.1.2
## [78] cachem_1.0.4
## [79] GenomicFeatures_1.42.2
## [80] fs_1.5.0
## [81] GetoptLong_1.0.5
## [82] clusterProfiler_4.7.1
## [83] DelayedMatrixStats_1.12.3
## [84] vctrs_0.6.2
## [85] generics_0.1.0
## [86] plot3D_1.4
## [87] tools_4.0.3
## [88] foreign_0.8-81
## [89] NOISeq_2.34.0
## [90] munsell_0.5.0
## [91] tweenr_1.0.2
## [92] fgsea_1.16.0
## [93] DelayedArray_0.16.3
## [94] fastmap_1.1.0
## [95] compiler_4.0.3
## [96] abind_1.4-5
## [97] rtracklayer_1.50.0
## [98] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
## [99] GenomeInfoDbData_1.2.4
## [100] gridExtra_2.3
## [101] edgeR_3.32.1
## [102] lattice_0.20-45
## [103] ggnewscale_0.4.7
## [104] AnnotationForge_1.32.0
## [105] utf8_1.2.1
## [106] dplyr_1.1.2
## [107] BiocFileCache_1.14.0
## [108] jsonlite_1.8.4
## [109] scales_1.2.1
## [110] graph_1.68.0
## [111] carData_3.0-4
## [112] sparseMatrixStats_1.2.1
## [113] TFEA.ChIP_1.10.0
## [114] genefilter_1.72.1
## [115] car_3.0-11
## [116] doParallel_1.0.16
## [117] latticeExtra_0.6-29
## [118] R.utils_2.12.0
## [119] brew_1.0-6
## [120] checkmate_2.0.0
## [121] rmarkdown_2.14
## [122] openxlsx_4.2.3
## [123] pkgdown_1.6.1
## [124] cowplot_1.1.1
## [125] textshaping_0.3.6
## [126] forcats_1.0.0
## [127] downloader_0.4
## [128] BSgenome_1.58.0
## [129] igraph_1.4.99.9024
## [130] survival_3.2-10
## [131] numDeriv_2016.8-1.1
## [132] yaml_2.2.1
## [133] plotrix_3.8-2
## [134] systemfonts_1.0.4
## [135] ashr_2.2-47
## [136] SQUAREM_2021.1
## [137] htmltools_0.5.2
## [138] memoise_2.0.0
## [139] VariantAnnotation_1.36.0
## [140] locfit_1.5-9.4
## [141] graphlayouts_0.7.1
## [142] batchtools_0.9.15
## [143] PCAtools_2.2.0
## [144] viridisLite_0.4.0
## [145] rrcov_1.7-0
## [146] digest_0.6.27
## [147] assertthat_0.2.1
## [148] rappdirs_0.3.3
## [149] emdbook_1.3.12
## [150] RSQLite_2.2.5
## [151] amap_0.8-18
## [152] yulab.utils_0.0.4
## [153] debugme_1.1.0
## [154] misc3d_0.9-1
## [155] data.table_1.14.2
## [156] blob_1.2.1
## [157] R.oo_1.24.0
## [158] ragg_0.4.0
## [159] labeling_0.4.2
## [160] splines_4.0.3
## [161] Cairo_1.5-12.2
## [162] ggupset_0.3.0
## [163] RCurl_1.98-1.3
## [164] broom_1.0.4
## [165] hms_1.1.3
## [166] colorspace_2.0-0
## [167] BiocManager_1.30.16
## [168] GenomicRanges_1.42.0
## [169] shape_1.4.6
## [170] sass_0.4.1
## [171] GEOquery_2.58.0
## [172] Rcpp_1.0.9
## [173] mvtnorm_1.1-2
## [174] circlize_0.4.15
## [175] enrichplot_1.10.2
## [176] fansi_0.4.2
## [177] tzdb_0.3.0
## [178] truncnorm_1.0-8
## [179] ChIPseeker_1.33.0.900
## [180] R6_2.5.0
## [181] grid_4.0.3
## [182] lifecycle_1.0.3
## [183] ShortRead_1.48.0
## [184] zip_2.1.1
## [185] curl_4.3
## [186] ggsignif_0.6.3
## [187] jquerylib_0.1.3
## [188] robustbase_0.95-0
## [189] DO.db_2.9
## [190] Matrix_1.5-4
## [191] qvalue_2.22.0
## [192] desc_1.3.0
## [193] org.Hs.eg.db_3.12.0
## [194] RColorBrewer_1.1-2
## [195] iterators_1.0.13
## [196] stringr_1.5.0
## [197] DOT_0.1
## [198] ggpie_0.2.5
## [199] beachmat_2.6.4
## [200] polyclip_1.10-0
## [201] biomaRt_2.46.3
## [202] purrr_1.0.1
## [203] shadowtext_0.0.9
## [204] gridGraphics_0.5-1
## [205] mgcv_1.8-34
## [206] ComplexHeatmap_2.13.1
## [207] openssl_1.4.3
## [208] patchwork_1.0.0
## [209] bdsmatrix_1.3-4
## [210] codetools_0.2-18
## [211] matrixStats_0.58.0
## [212] invgamma_1.1
## [213] GO.db_3.12.1
## [214] gtools_3.8.2
## [215] prettyunits_1.1.1
## [216] dbplyr_2.3.2
## [217] R.methodsS3_1.8.1
## [218] GenomeInfoDb_1.26.7
## [219] gtable_0.3.0
## [220] DBI_1.1.1
## [221] highr_0.8
## [222] ggfun_0.0.6
## [223] httr_1.4.5
## [224] KernSmooth_2.23-18
## [225] stringi_1.5.3
## [226] progress_1.2.2
## [227] reshape2_1.4.4
## [228] farver_2.1.0
## [229] annotate_1.68.0
## [230] viridis_0.6.1
## [231] Rgraphviz_2.34.0
## [232] xml2_1.3.4
## [233] bbmle_1.0.24
## [234] systemPipeR_1.24.3
## [235] boot_1.3-28
## [236] readr_2.1.4
## [237] geneplotter_1.68.0
## [238] ggplotify_0.1.0
## [239] Category_2.56.0
## [240] DEoptimR_1.0-11
## [241] DESeq2_1.30.1
## [242] bit_4.0.4
## [243] scatterpie_0.1.7
## [244] jpeg_0.1-8.1
## [245] MatrixGenerics_1.2.1
## [246] ggraph_2.0.5
## [247] pkgconfig_2.0.3
## [248] rstatix_0.7.0
## [249] knitr_1.37